| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,511,885 – 15,511,980 |
| Length | 95 |
| Max. P | 0.966842 |

| Location | 15,511,885 – 15,511,980 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.21 |
| Shannon entropy | 0.40724 |
| G+C content | 0.46934 |
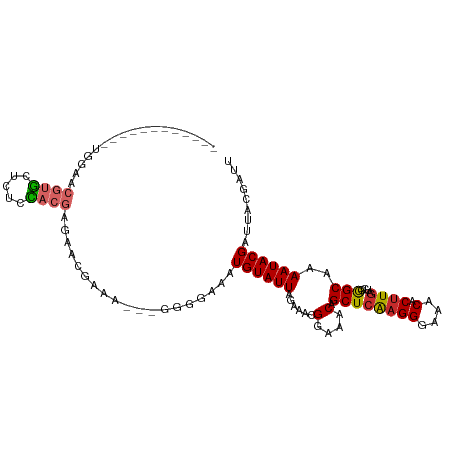
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.24 |
| Covariance contribution | -0.02 |
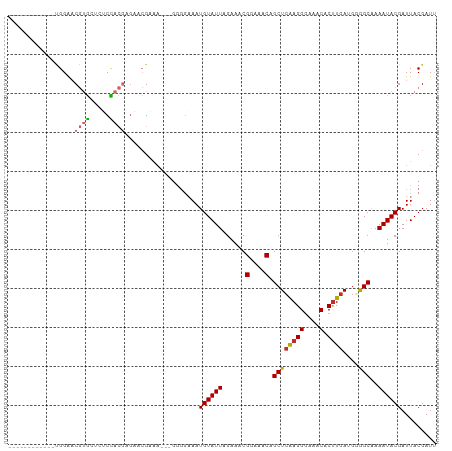
| Combinations/Pair | 1.21 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
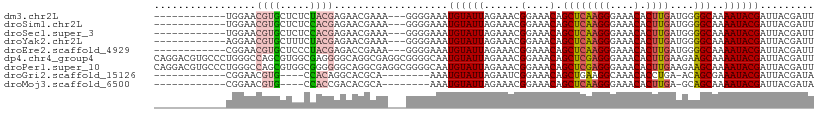
>dm3.chr2L 15511885 95 - 23011544 ------------UGGAACGUGCUCUCUACGAGAACGAAA---GGGGAAAUGUAUUAGAAACGGAAACAGCUCAAGGGAAACACUUGAUGGGGCAAAAUACGAUUACGAUU ------------.....((((.((..........(....---).......(((((......(....).((((((((....).)))))....))..))))))).))))... ( -19.10, z-score = -1.92, R) >droSim1.chr2L 15269225 95 - 22036055 ------------UGGAACGUGCUCUCCACGAGAACGAAA---GGGGAAAUGUAUUAGAAACGGAAACAGCUCAAGGGAAACACUUGAUGGGGCAAAAUACGAUUACGAUU ------------.....((((.(((((.((....))...---)))))..((((((......(....).((((((((....).)))))....))..))))))..))))... ( -20.90, z-score = -2.03, R) >droSec1.super_3 1764817 95 - 7220098 ------------UGGAACGUGCUCUCCACGAGAACGAAA---GGGGAAAUGUAUUAGAAACGGAAACAGCUCAAGGGAAACACUUGAUGGGGCAAAAUACGAUUACGAUU ------------.....((((.(((((.((....))...---)))))..((((((......(....).((((((((....).)))))....))..))))))..))))... ( -20.90, z-score = -2.03, R) >droYak2.chr2L 2506689 95 + 22324452 ------------AGGAACGUGCUUUCUACGAGAACGAAA---GGGGAAAUGUAUUAGAAACGGAAACAGCUCAAGGGAAACACUUGAUGGGGCAAAAUACGAUUACGAUU ------------.....(((((((((.........))))---)......((((((......(....).((((((((....).)))))....))..))))))..))))... ( -18.80, z-score = -2.12, R) >droEre2.scaffold_4929 7257622 95 - 26641161 ------------CGGAACGUGCUCCCUACGAGACCGAAA---GGGGAAAUGUAUUAGAAACGGAAACAGCUCAAGGGAAACACUUGAUGGGGCAAAAUACGAUUACGAUU ------------((.....((((((.((((...((....---)).....))))........(....)...((((((....).))))).)))))).....))......... ( -24.60, z-score = -2.63, R) >dp4.chr4_group4 4291128 110 + 6586962 CAGGACGUGCCCUGGGCCAGCGUGGCGAGGGGCAGGCGAGGCGGGGCAAUGUAUUAGAAACGGAAACAGCUCGAGGGAAACACUUGAAGAAGCAAAAUACGAUUACGAUU .....((((((((..(((.((.((.(....).)).))..))))))))..((((((......(....).((((((((....).)))))....))..))))))...)))... ( -35.70, z-score = -2.74, R) >droPer1.super_10 2127588 110 + 3432795 CAGGACGUGCCCUGGGCCAGCGUGGCGGGGGGCAGGCGAGGCGGGGCAAUGUAUUAGAAACGGAAACAGCUCGAGGGAAACACUUGAAGAAGCAAAAUACGAUUACGAUU .....((((((((..(((.((.((.(....).)).))..))))))))..((((((......(....).((((((((....).)))))....))..))))))...)))... ( -35.70, z-score = -2.62, R) >droGri2.scaffold_15126 5718837 85 - 8399593 ------------CGGAACGUG----CCACAGGCACGCA--------AAAUGUAUUAGAAUCGGAAACAGCUGAAGGCAAACACCUGA-ACAGCGAAAUACGAUUACGAUA ------------.....((((----((...))))))..--------.....((((..(((((......((((.(((......)))..-.))))......)))))..)))) ( -23.00, z-score = -3.54, R) >droMoj3.scaffold_6500 20911265 85 - 32352404 ------------CGGAACGUG----CCACCGACACGCA--------AAAUGUAUUAGAAACGGAAACAGCUCAAGGGAAACACUUGA-GCAGCAAAAUACGAUUACGAUA ------------((...((((----.......))))..--------...((((((......(....).((((((((....).)))))-)).....))))))....))... ( -22.90, z-score = -4.17, R) >consensus ____________UGGAACGUGCUCUCCACGAGAACGAAA___GGGGAAAUGUAUUAGAAACGGAAACAGCUCAAGGGAAACACUUGAUGGGGCAAAAUACGAUUACGAUU .................((((.....))))...................((((((......(....).((((((((....).))))....)))..))))))......... (-12.27 = -12.24 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:00 2011