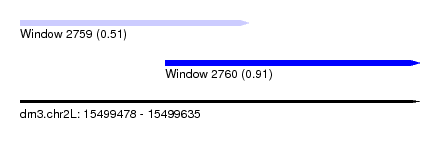
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,499,478 – 15,499,635 |
| Length | 157 |
| Max. P | 0.906454 |

| Location | 15,499,478 – 15,499,568 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 63.93 |
| Shannon entropy | 0.65368 |
| G+C content | 0.42995 |
| Mean single sequence MFE | -19.81 |
| Consensus MFE | -5.21 |
| Energy contribution | -5.69 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15499478 90 + 23011544 GACGAAACGAAAAUAACGAGCU-AAAAGGCCAUAAAAAUUGGCCAAAAAU---GUGGUUCCAUA-----GCUGGUUCCAUUCACCCUCUCGCUCGCUCU ................(((((.-....(((((.......)))))......---((((..(((..-----..)))..))))..........))))).... ( -24.50, z-score = -3.18, R) >droEre2.scaffold_4929 7245154 90 + 26641161 UCCGAAACCAAAAUAACGAGCC-AAAGAGCCAUAAAAAUUGGCCAAAAAUCGCGUGGUGCCACU-----GCUCG---CGCUCUCCCUCUCGCUCGCUCC ................(((((.-..(((((((.......))))........((((((.((....-----)))))---)))......))).))))).... ( -22.70, z-score = -2.47, R) >droYak2.chr2L 2493900 87 - 22324452 CCCGAAACCAAAAUAACGAGCC-AAAAAGCCAUAAAAAUUGGCCUAAAAU---GUGGUUCCAUU-----GCUCG---CGCUCACCCUCUCGCUCGCUCU ................(((((.-.....((((.......)))).......---(((((......-----)).))---)............))))).... ( -15.70, z-score = -1.31, R) >droSec1.super_3 1752319 87 + 7220098 GCCGAAACCAAAAUAACGAGCU-AUAAAGCCAUAAAAAUUGGCCAAAAAU---AUGGUUCCAUA-----CCUGG---CACACAUUCUCUCGCUCGCUCU ................(((((.-.....((((.......)))).......---(((((.(((..-----..)))---.)).)))......))))).... ( -16.80, z-score = -2.08, R) >droSim1.chr2L 15256770 87 + 22036055 GCCGAAACCAAAAUAUCGAGCU-AUAAAGCCAUAAAAAUUGGCCAAAAAU---GUGGUUCCAUA-----GCUGG---CACACACCCUCUCGCUCGCUCU ................(((((.-.....((((.......)))).......---(((((.(((..-----..)))---.)).)))......))))).... ( -18.50, z-score = -1.57, R) >droVir3.scaffold_12963 12178254 90 + 20206255 -AAAGAAAGAAAAAAAAAACCACAACUAACAGA-AAGGCGAGGCAA-UUUCGAAUAUGUUUUU------GCUUUUGCCUCUGUUUCUGCUUCUUGUUCU -....................((((..(.((((-((.(.(((((((-...((((......)))------)...)))))))).)))))).)..))))... ( -21.00, z-score = -1.81, R) >droMoj3.scaffold_6500 20895411 97 + 32352404 -AAAAAACGAAAAUAAAAAAGA-AACAAGCGGGCAAGACAAGGCAAAUUUGGAAUAUUUUUUUUUUCACGCUUUUGCUACAGUUUCUGCUUCUUGUUCU -..................(((-(.((((.(((((((((..((((((..(((((..........))).))..))))))...)))).))))))))))))) ( -19.50, z-score = -0.96, R) >consensus GCCGAAACCAAAAUAACGAGCU_AAAAAGCCAUAAAAAUUGGCCAAAAAU___GUGGUUCCAUA_____GCUGG___CACUCACCCUCUCGCUCGCUCU ................(((((.......((((.......))))..........(((....)))...........................))))).... ( -5.21 = -5.69 + 0.47)
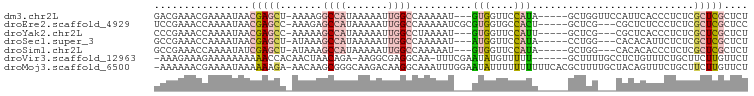
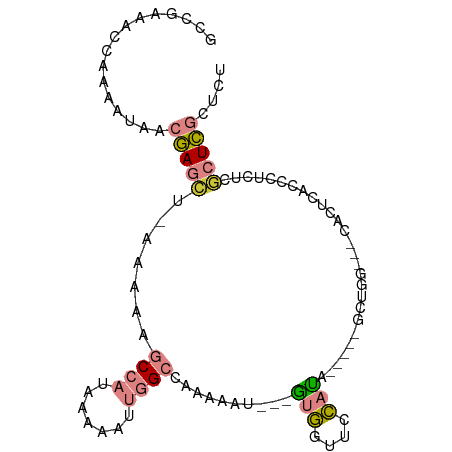
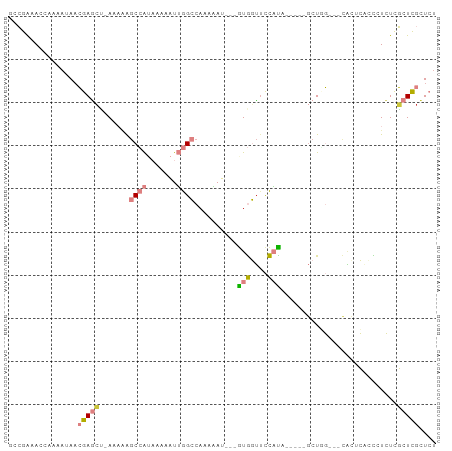
| Location | 15,499,535 – 15,499,635 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 60.77 |
| Shannon entropy | 0.61895 |
| G+C content | 0.55776 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -12.80 |
| Energy contribution | -13.42 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15499535 100 + 23011544 AUAGCUGGUUCCAUUCACCCUCUCGCUCGCUCUUCCAGCCGGCGAAUUAUUGAUUUUGUGUGAGAGCGAGGGAGCGCGCAUAAGAGAGAGGGAGAGAGAG .............(((.((((((((((.(((.....))).)))..........(((((((((...((......)).)))))))))))))))).))).... ( -34.00, z-score = -0.54, R) >droEre2.scaffold_4929 7245214 82 + 26641161 ---ACUGCUCGCGCUCUCCCUCUCGCUCGCUCCUCCAGCCGGCGAACCAUUGAUUUUGCGCGAGAGCGUGUGAGUG----CGAGAGAGA----------- ---.((.(((((((((.(((((((((..(((.....)))..(((((........)))))))))))).).).)))))----))))))...----------- ( -36.40, z-score = -2.32, R) >droYak2.chr2L 2493957 83 - 22324452 ---AUUGCUCGCGCUCACCCUCUCGCUCGCUCUUCCAGCCGGCGAACCAUUCAUUUUGUGGGAGAGCGAGCGAGAG----UGAGAGAGAG---------- ---....(((...(((((..((((((((((((((((.....(((((........))))))))))))))))))))))----)))).)))..---------- ( -42.70, z-score = -4.23, R) >droMoj3.scaffold_6500 20895477 94 + 32352404 CGCUUUUGCUACAGUUUCUGCUUCUUGUUCUCCUCGUGCCGGUGCUUCAUUGAUUUUGCUUGCAAGUAUGUGUGUG----UGCGAGAGAGCAAGUCCA-- .((....((....))....))..(((((((((.(((..(((((((((((...........)).))))))...)).)----..))))))))))))....-- ( -25.50, z-score = -0.69, R) >consensus ___ACUGCUCGCACUCACCCUCUCGCUCGCUCCUCCAGCCGGCGAACCAUUGAUUUUGCGUGAGAGCGAGUGAGUG____UGAGAGAGAG__________ .....................(((((((((((((.(.....(((((........)))))).))))))))))))).......................... (-12.80 = -13.42 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:59 2011