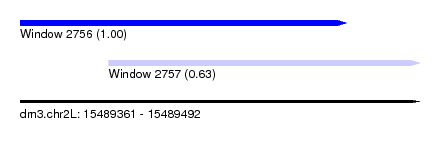
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,489,361 – 15,489,492 |
| Length | 131 |
| Max. P | 0.997708 |

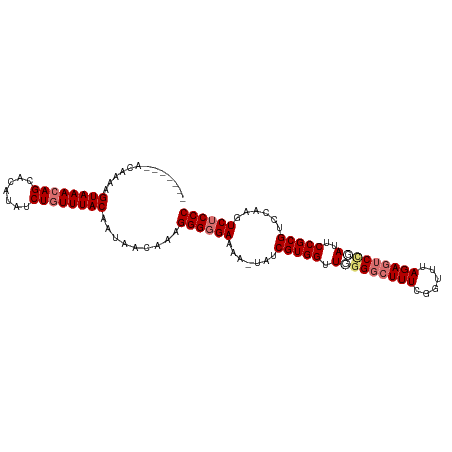
| Location | 15,489,361 – 15,489,468 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Shannon entropy | 0.15226 |
| G+C content | 0.43255 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -27.07 |
| Energy contribution | -28.57 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15489361 107 + 23011544 AAAAAAAAGAAAAGUAAACAGCACAUAUCUGUUUACAAUAACAAAGGGGGAAAAAUAUCGUGGUUGGGGCUUUCGGUUUAGAGUCCGAUUCCGCGUCCAAGUCUCCC .............((((((((.......)))))))).........((((((.......(((((.(.(((((((......))))))).)..)))))......)))))) ( -33.32, z-score = -4.07, R) >droSim1.chr2L 15243172 99 + 22036055 -------ACAAAAGUAAACAGCACAUAUCUGUUUACAAUAACAAAGGGGGAAAA-UAUCGUGGUUGGGGCUUUCGGUUUAGAGUCUCAUACCGCGUCCAAGUCUCCC -------......((((((((.......)))))))).........((((((...-...(((((((((((((((......))))))))).))))))......)))))) ( -36.60, z-score = -5.81, R) >droYak2.chr2L 2482821 100 - 22324452 -------ACAAAAGUAAACAGCACAUAUCUAUUUACAAUAACAAAGGGGGAAAAAUAUCGUGGUUGGGCAUUUCGGUUUAGAGUCGGAUUCCGCGUCCAAGUCUCCC -------......(((((.((.......)).))))).........((((((.......(((((((.(((...((......))))).))..)))))......)))))) ( -20.92, z-score = -0.83, R) >droEre2.scaffold_4929 7235317 99 + 26641161 -------ACAAAAGUAAACAGCACAUAUCUGUUUACAAUAACAAAGGGGGAAAA-CGUCGUGGUUUGGGCUUUCGGUUUAGAGUCCGAUUCCGCGUCCAAGUCGCCC -------......((((((((.......)))))))).........(((.((...-.(.(((((.(((((((((......)))))))))..))))).)....)).))) ( -34.10, z-score = -3.91, R) >consensus _______ACAAAAGUAAACAGCACAUAUCUGUUUACAAUAACAAAGGGGGAAAA_UAUCGUGGUUGGGGCUUUCGGUUUAGAGUCCGAUUCCGCGUCCAAGUCUCCC .............((((((((.......)))))))).........((((((.......(((((.(((((((((......)))))))))..)))))......)))))) (-27.07 = -28.57 + 1.50)

| Location | 15,489,390 – 15,489,492 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.27 |
| Shannon entropy | 0.18140 |
| G+C content | 0.46124 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -22.25 |
| Energy contribution | -23.25 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15489390 102 + 23011544 UGUUUACAAUAACAAAGGGGGAAAAAUAUCGUGGUUGGGGCUUUCGGUUUAGAGUCCGAUUCCGCGUCCAAGUCUCCCCAACUACUCGUA---UCCGUACAAUAG ((((......))))..((((((.......(((((.(.(((((((......))))))).)..)))))......)).))))........(((---....)))..... ( -27.42, z-score = -1.23, R) >droSim1.chr2L 15243194 104 + 22036055 UGUUUACAAUAACAAAGGGGGAAAA-UAUCGUGGUUGGGGCUUUCGGUUUAGAGUCUCAUACCGCGUCCAAGUCUCCCCAACUACUCGUACAAUCCGUACAAUAG ((((......))))..((((((...-...(((((((((((((((......))))))))).))))))......)).))))........((((.....))))..... ( -33.80, z-score = -3.71, R) >droYak2.chr2L 2482843 96 - 22324452 UAUUUACAAUAACAAAGGGGGAAAAAUAUCGUGGUUGGGCAUUUCGGUUUAGAGUCGGAUUCCGCGUCCAAGUCUCCCCAACUA---------UCCGUACAAUAG ................((((((.......(((((((.(((...((......))))).))..)))))......)).)))).....---------............ ( -19.62, z-score = 0.28, R) >droEre2.scaffold_4929 7235339 95 + 26641161 UGUUUACAAUAACAAAGGGGGAAAA-CGUCGUGGUUUGGGCUUUCGGUUUAGAGUCCGAUUCCGCGUCCAAGUCGCCCCAACUA---------UCCGCACAAUAG ((((......))))..((((((...-.(.(((((.(((((((((......)))))))))..))))).)....)).)))).....---------............ ( -29.60, z-score = -2.03, R) >consensus UGUUUACAAUAACAAAGGGGGAAAA_UAUCGUGGUUGGGGCUUUCGGUUUAGAGUCCGAUUCCGCGUCCAAGUCUCCCCAACUA_________UCCGUACAAUAG ................((((((.......(((((.(((((((((......)))))))))..)))))......)).)))).......................... (-22.25 = -23.25 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:57 2011