
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,471,594 – 15,471,701 |
| Length | 107 |
| Max. P | 0.695449 |

| Location | 15,471,594 – 15,471,701 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Shannon entropy | 0.48021 |
| G+C content | 0.46432 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -9.29 |
| Energy contribution | -9.75 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15471594 107 - 23011544 UUACCUGUUCUGAUUUGAACGCUGAAAGUGAAACCGUAAAUUCCAUUC-----CCGAGGGCCCAAUGCGAGUCGUUUACCUUU-UGGCCAUCAAAUCAGACAUCAGGUGGCAG .((((((.((((((((((..((..((((..((((.....(((((((((-----(....))...)))).)))).))))..))))-..))..))))))))))...)))))).... ( -36.20, z-score = -3.78, R) >droSim1.chr2L 15224395 107 - 22036055 UUACCUGUUCUGAUUUGAACGCUGAAAGUGAAACCGUAAAUUCCAUUC-----CCGAGGGCCCAAUGCGAGUCGUUUUCCUUU-UGGCCAUCAAAUCAGACAUCAGGUGGCAG .((((((.((((((((((..((..((((.((((.((...(((((((((-----(....))...)))).)))))).))))))))-..))..))))))))))...)))))).... ( -39.10, z-score = -4.58, R) >droSec1.super_3 1723950 107 - 7220098 UUACCUGUUCUGAUUUGAACGCUGAAAGUGAAACCGUAAAUUCCAUUC-----CCGAGGGCCCAAUGCCAGUCGUUUUCCUUU-UGGCCAUCAAAUCAGACAUCAGGUGGCAG .((((((.((((((((((..((..((((..((((((..(((...))).-----.))..(((.....)))....))))..))))-..))..))))))))))...)))))).... ( -37.10, z-score = -4.20, R) >droYak2.chr2L 2463561 107 + 22324452 UUACCUGUUCUGAUUUGAACGCUGAAAGUGAAACCGUAAAUUCCACUC-----CCGAGGGCCCAAUGAGAGUCGUGUUCCUUU-UGGCCAUCAAAUCAUACAUCAGGUGGCAG .((((((...((((((((..((..((((.(((..((...((((((..(-----(....)).....)).))))))..)))))))-..))..)))))))).....)))))).... ( -30.80, z-score = -2.03, R) >droEre2.scaffold_4929 7217703 107 - 26641161 UUACCUGUUCUGAUUUGAACGCUGAAAGUGAAACAGUAAAUUGCAUUC-----CCGAGGGCCCAAUGCGAGUCGUUUUCCUCU-UGGCCAUCAAAUCAGACAUCAGGUGGCAG .((((((.((((((((((..((..(.((..((((.(....((((((((-----(....))...)))))))..)))))..)).)-..))..))))))))))...)))))).... ( -38.10, z-score = -3.83, R) >droAna3.scaffold_12916 7776658 101 - 16180835 UUACCUGUUCUGAUUUGAAGACUGAAAGUGAAACCGUAAAUUUAAUUC-----CCGAGGGUCCAAUGCCCGUCUUCUACU-------ACAUCAAAUCAGACAUCAGGUGGCAG .((((((.((((((((((((((............((..(((...))).-----.)).((((.....))))))))......-------...))))))))))...)))))).... ( -27.30, z-score = -2.44, R) >dp4.chr4_group4 4243496 100 + 6586962 UUACCUGUUCUGAUUUGAGCGCUGAAAGUGAAACCGUAAAU-----CC-----UCGAGGGUCUAAUUUGAGUCGAGUCUU--U-GGGCUAUCAAAUCAAACAUCAGGUGUCGG .((((((...((((((((..(((.(((((((.....((.((-----((-----....)))).)).......)))...)))--)-.)))..)))))))).....)))))).... ( -26.70, z-score = -1.25, R) >droPer1.super_10 2081022 100 + 3432795 UUACCUGUUCUGAUUUGAGCGCUGAAAGUGAAACCGUAAAU-----CC-----UCGAGGGUCUAAUUUGAGUCGAGUCUU--U-GGGCUAUCAAAUCAAACAUCAGGUGUCGG .((((((...((((((((..(((.(((((((.....((.((-----((-----....)))).)).......)))...)))--)-.)))..)))))))).....)))))).... ( -26.70, z-score = -1.25, R) >droGri2.scaffold_15126 5673288 99 - 8399593 CUACCUGUUCAGAUUUCAGAGCUGAAAGUGAAACCAUAAAU-------------UGAGUCUCGUAGGCAGUGGGCAUCGCUUU-UGGCUAUCAAAUCAAACAUCAGGUGUCGG .((((((....(((((...(((..(((((((..((((....-------------...(((.....))).))))...)))))))-..)))...)))))......)))))).... ( -29.30, z-score = -2.35, R) >droMoj3.scaffold_6500 20860241 112 - 32352404 -UACCUGUUCUGAUUUCAGCGCUGAAAGUGAAACCGUAAAUUCUCGGCUUGGGCUCGGGCUCAGGCCCAGGUCCGAUCGCCUUGCGGCUAUCAAAUCAAACAUCAGGUGUCGG -((((((...((((((....((((((.((((..(((........))).((((((..((((....))))..)))))))))).)).))))....)))))).....)))))).... ( -39.50, z-score = -2.17, R) >droVir3.scaffold_12963 12147398 101 - 20206255 -UACCUGUUCUGAUUUCAGCGCUGAAAGUGAAACCGUAAAUUCCAG--------UCAGUCGCAGAGCCCGAGC--AUCGCCUU-UGGCUAUCAAAUCAAACAUCAGGUGUCGG -((((((...((((((..(((((((..(.(((........))))..--------)))).)))..((((.(((.--.....)))-.))))...)))))).....)))))).... ( -27.80, z-score = -1.95, R) >consensus UUACCUGUUCUGAUUUGAACGCUGAAAGUGAAACCGUAAAUUCCAUUC_____CCGAGGGCCCAAUGCGAGUCGUUUCCCUUU_UGGCCAUCAAAUCAAACAUCAGGUGGCAG .((((((...((((((...(((.....)))...((......................)).................................)))))).....)))))).... ( -9.29 = -9.75 + 0.46)

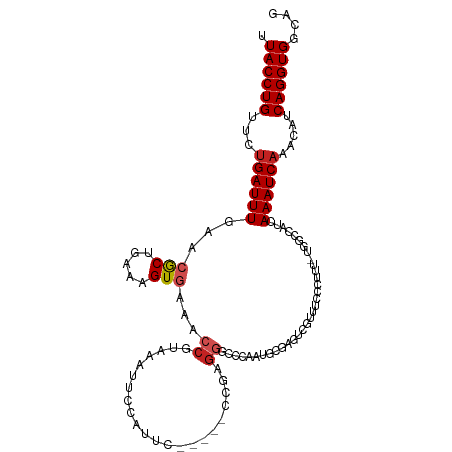
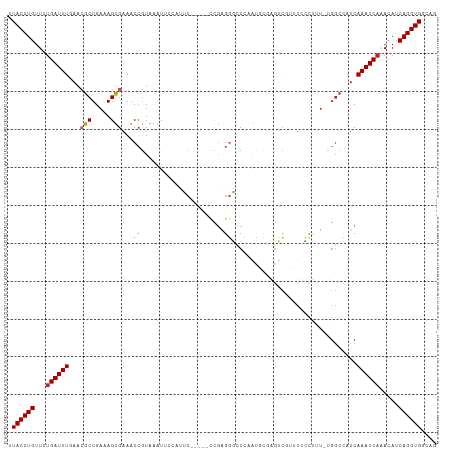
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:54 2011