
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,427,965 – 15,428,081 |
| Length | 116 |
| Max. P | 0.741076 |

| Location | 15,427,965 – 15,428,081 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Shannon entropy | 0.39537 |
| G+C content | 0.49773 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -19.93 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.741076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15427965 116 + 23011544 CGCACAGAGUCGCAUAAUGAAGUGCAAAAUGUCAUUAAUUUGCCGCCGUCAAGGUGGCAAGGCAGCGAGUAUCUGCGGAUCGGGGCAUCCAACUUCAAACUAUCCGAGUAUCUGUG ..((((((.(((.(((.((((((.....(((((.....(((((((((.....)))))))))((((.......)))).......)))))...))))))...))).)))...)))))) ( -39.10, z-score = -2.45, R) >droSim1.chr2L 15187824 101 + 22036055 CGCACAGAGUCGCAUAAUGAAGUGCAAAAUGUCAUUAAUUUGCCGCCGUCAAGGUGGCAAG------GCAGC--GAGUAUCUGCGGAUCGGGGC-------AUCCAACUAUCUGUG ..((((((.((((.((((((.((.....)).)))))).(((((((((.....)))))))))------...))--))........((((......-------)))).....)))))) ( -35.20, z-score = -2.19, R) >droSec1.super_3 1687961 101 + 7220098 CGCACAGAGUCGCAUAAUGAAGUGCAAAAUGUCAUUAAUUUGCCGCCGUCAAGGUGGCAAG------GCAGC--GAGUAUCUGCGUAUCGGGGC-------AUCCAACUAUCUGUG ..((((((...((((......))))...(((((.....(((((((((.....)))))))))------((((.--......)))).......)))-------)).......)))))) ( -33.90, z-score = -2.07, R) >droYak2.chr2L 2426903 101 - 22324452 CGCACAGAGUCGCAUAAUGAAGUGCAAAAUGCCAUUAAUUUGCCGCCGUCAAGGUGGCAAG--------UAUCUGGGGAUUGGGGCAUCCAACC-------AUCCGAGUAUCUGGG ....((((.((((((......))))......(((...((((((((((.....)))))))))--------)...)))((((.((.........))-------))))))...)))).. ( -33.90, z-score = -1.55, R) >droEre2.scaffold_4929 7178052 106 + 26641161 CGCACAGAGUCGCAUAAUGAAGUGCAAAAUGCCAUUAAUUUGCCGCCGUCAAGGUGGCAAGGCAAC---CAUCCAACCAUCCAAACAGCCGAGU-------AUCUGGGUAUCUGGG ....((((...((((......))))....((((......((((((((.....))))))))))))..---..................(((.((.-------..)).))).)))).. ( -29.70, z-score = -0.78, R) >droAna3.scaffold_12916 7734871 104 + 16180835 CGCACAGAGUCGCAUAAUGAAGUGCAAAAUGUCAUUAAUUUGCCGCCGUCAAGGUGGCAUUU-----GCUGCAAGAGUAUCUGAGUUUCUAAGU-------AUCUGAGUAACCGAG .((.((((((.(((((((((.((.....)).))))))...(((((((.....)))))))..)-----)).)).((((.........))))....-------.)))).))....... ( -26.80, z-score = -0.99, R) >consensus CGCACAGAGUCGCAUAAUGAAGUGCAAAAUGUCAUUAAUUUGCCGCCGUCAAGGUGGCAAG______GCUACC_GAGUAUCUGAGCAUCCAAGC_______AUCCGAGUAUCUGUG ....((((...((((......)))).............(((((((((.....))))))))).................................................)))).. (-19.93 = -20.60 + 0.67)

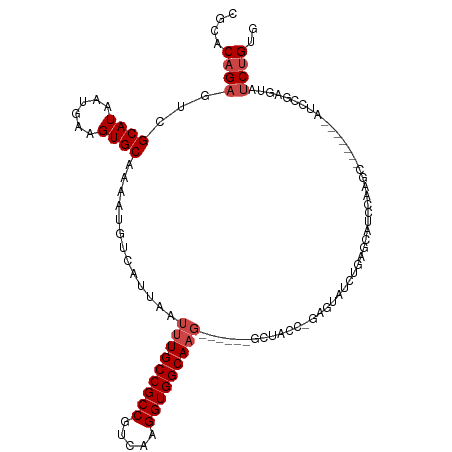
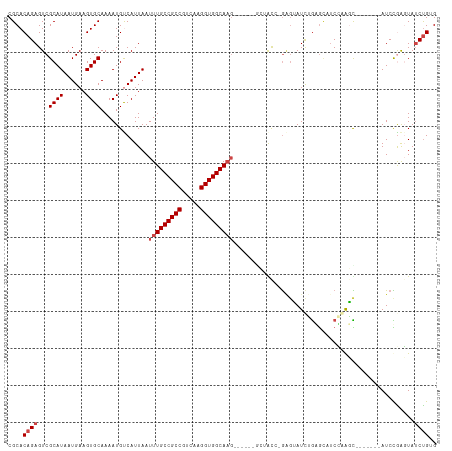
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:52 2011