| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,421,112 – 1,421,205 |
| Length | 93 |
| Max. P | 0.887276 |

| Location | 1,421,112 – 1,421,205 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 60.74 |
| Shannon entropy | 0.79162 |
| G+C content | 0.51444 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -5.88 |
| Energy contribution | -5.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
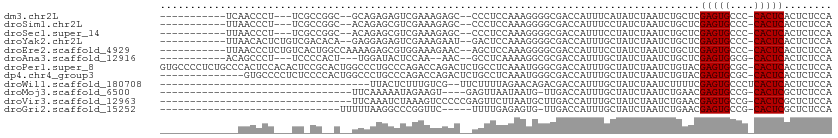
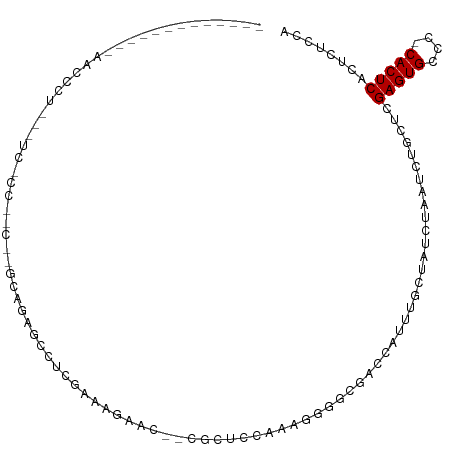
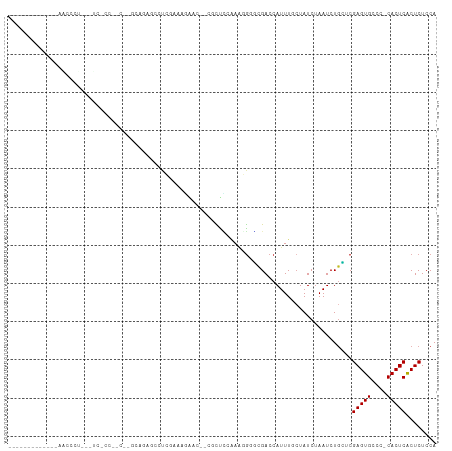
>dm3.chr2L 1421112 93 + 23011544 -----------UCAACCCU---UCGCCGGC--GCAGAGAGUCGAAAGAGC--CCCUCCAAAGGGGCGACCAUUUCAUAUCUAAUCUGCUCGAGUGCCC-CACUCACUCUCCA -----------........---.....((.--(((((...((....))((--((((....)))))).................)))))..(((((...-))))).....)). ( -28.20, z-score = -2.24, R) >droSim1.chr2L 1394275 93 + 22036055 -----------UUAACCCU---UCGCCGGC--ACAGAGCGUCGAAAGAGC--CCCUCCAAAGGGGCGACCAUUUCCUAUCUAAUCUGCUCGAGUGCCC-CACUCACUCUCCA -----------........---.....(((--((.((((.((....))((--((((....))))))....................))))..))))).-............. ( -30.50, z-score = -3.60, R) >droSec1.super_14 1368666 93 + 2068291 -----------UUAACCCU---UCGCCGGC--ACAGAGCGUCGAAAGAGC--CCCUCCAAAGGGGCGACCAUUUCCUAUCUAAUCUGCUCGAGUGCCC-CACUCACUCUCCA -----------........---.....(((--((.((((.((....))((--((((....))))))....................))))..))))).-............. ( -30.50, z-score = -3.60, R) >droYak2.chr2L 1395944 96 + 22324452 -----------UUAACACUCUGUCGACACA--GAGGAGAGUCGAAAGAAU--GACUCCAAAGGGGCGACCAUUUGCUAUCUAAUCUGCUCGAGUGCCC-CACUCACUCUCCA -----------.......(((((....)))--))((((((((....))..--.........((((((.......((..........)).....)))))-).....)))))). ( -29.80, z-score = -2.35, R) >droEre2.scaffold_4929 1464006 98 + 26641161 -----------UUAACCCUCUGUCACUGGCCAAAAGAGCGUGGAAAGAAC--AGCUCCAAAGGGGCGACCAUUUCCUAUCUAAUCUGCUCGAGUGCCC-CACUCACUCUCCA -----------................((......(((((.(((((....--.(((((...))))).....))))).........)))))(((((...-))))).....)). ( -22.80, z-score = -0.27, R) >droAna3.scaffold_12916 4883411 90 + 16180835 -----------ACAGCCCU---UCCCCACU---UGGAUACUCCAA--AAC--GCCUCAAAAGGCGCGACCAUUUGCUAUCUAAUCUGCUCGAGUGGCG-CACUCACUCUCCA -----------........---.......(---(((.....))))--...--((((....))))(((.((((((((..........))..))))))))-)............ ( -20.30, z-score = -1.85, R) >droPer1.super_8 1151650 111 + 3966273 GUGCCCCUCUGCCCACUCCACACUCCGCACUGGCCCUGCCCAGACCAGACUCUGCCUCAAAUGGGCGACCAUUUGCUAUCUAAUCUGUACGAGUGCGC-CACUCACUCUCCA (((.......((.(((((...........((((......))))..((((........(((((((....)))))))........))))...))))).))-....)))...... ( -26.49, z-score = -1.60, R) >dp4.chr4_group3 9944398 97 + 11692001 --------------GUGCCCCUCUCCCCACUGGCCCUGCCCAGACCAGACUCUGCCUCAAAUGGGCGACCAUUUGCUAUCUAAUCUGUACGAGUGCGC-CACUCACUCUCCA --------------((((..(((......((((......))))..((((........(((((((....)))))))........))))...))).))))-............. ( -24.19, z-score = -1.90, R) >droWil1.scaffold_180708 3101695 75 + 12563649 -----------------------------------UUACUCUUUGUCG--UUCUUUUAGAACAGACGACCAUUUGCUAUCUAAUCUUUUCGAGUGCCCUCACUCACUCUCCA -----------------------------------.......((((((--(((.....)))).)))))......................(((((....)))))........ ( -13.40, z-score = -2.70, R) >droMoj3.scaffold_6500 28880754 74 - 32352404 --------------------------------UUCAAAAAUAGAAGU----GAGUUAAUAAUG-UUGACCAUUUGCUAUCUAAUCUGAACGAGUGCCG-CACUCGCUCUCCA --------------------------------.......((((((((----(.((((((...)-)))))))))).)))).......((.((((((...-)))))).)).... ( -16.10, z-score = -2.17, R) >droVir3.scaffold_12963 7398402 79 + 20206255 --------------------------------UUCAAAUCUAAAGUCCCCCGAGUUCUUAAUGCUUGACCAUUUGCUAUCUAAUCUGAACGAGUGCCG-CACUCGCUCUCCA --------------------------------..................(((((.......)))))...................((.((((((...-)))))).)).... ( -11.60, z-score = -1.01, R) >droGri2.scaffold_15252 10529119 75 + 17193109 ------------------------------UUUUUAAGGCCCGGUUC-----UUUUGAGAGUG-UUGACCAUUUGCUAUCUAAUCUGAACGAGUGCCG-CACUCGCUCUCCA ------------------------------.................-----....(((((((-.((..((((((...((......)).))))))...-))..))))))).. ( -14.90, z-score = -0.00, R) >consensus _____________AACCCU___UC_CC__C__GCAGAGCCUCGAAAGAAC__CGCUCCAAAGGGGCGACCAUUUGCUAUCUAAUCUGCUCGAGUGCCC_CACUCACUCUCCA ..........................................................................................(((((....)))))........ ( -5.88 = -5.88 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:32 2011