| Sequence ID | dm3.chr2L |
|---|---|
| Location | 127,044 – 127,138 |
| Length | 94 |
| Max. P | 0.519754 |

| Location | 127,044 – 127,138 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.32 |
| Shannon entropy | 0.51051 |
| G+C content | 0.49045 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.66 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 127044 94 + 23011544 --AAUACGUAGACUUU----AAAAGCUGGUAAUGAAUUC---CAGCUGCCAGGCAGCGACUAGCA------A----GUCGUAACCACUAG--GUACGCAAUGU--CUCUCAUUGAAC--- --..(((.(((.....----...((((((.........)---)))))....((..((((((....------)----)))))..)).))).--)))..(((((.--....)))))...--- ( -27.00, z-score = -2.28, R) >droSim1.chr2L 127832 100 + 22036055 --AAUACGUAGACUUU----AAAAGCUGGCAGUGAACCC---CAGCUGCCAGGCAGCGACUAGCAAGUCGUA----GUCGUAAUAACUAG--GUACGCAACGU--CUCUCAUUGAAC--- --..(((.(((.....----......(((((((......---..)))))))(((.((((((....)))))).----))).......))).--)))........--............--- ( -27.60, z-score = -1.21, R) >droSec1.super_14 125997 100 + 2068291 --AAUACGUAGACUUU----AAAAGCUGGCAGUGAACCC---CAGCUGCCAGGCAGCGACUAGCAAGUCGUA----GUCGUAAUAACUAG--GUACGCAACGU--CUCUCAUUGAAC--- --..(((.(((.....----......(((((((......---..)))))))(((.((((((....)))))).----))).......))).--)))........--............--- ( -27.60, z-score = -1.21, R) >droYak2.chr2L 116095 89 + 22324452 --AAUGCGUAGUCUUU----AAAAGCUGGCAGUGAACCC---CAGCUGCCAGGCAGCGACUAGCA-GUCGUA----GUCGUAACCACUAG--AUACGCAACGU--CU------------- --..((((((.(((..----.....((((((((......---..))))))))...(((((((...-....))----))))).......))--)))))))....--..------------- ( -31.00, z-score = -2.76, R) >droEre2.scaffold_4929 156718 90 + 26641161 --AAUACGUAGACUUU----AAAAGCUGGCAGUGAACCC---CAGCUGCCAGGCAGCGACUAGCAAGUCGUA----GUCGUAACCACUAG--GUAUGCAACGU--CU------------- --...((((.((((..----.....((((((((......---..))))))))...((((((....)))))))----)))((((((....)--)).))).))))--..------------- ( -31.60, z-score = -2.92, R) >droAna3.scaffold_12916 14198753 101 - 16180835 --AAUAAGCCUCCGCUGGCAAUAAGCUGGCAGUGAACCUUAGCAGCUGUCAGCCUGCGACUAGCUA----------GUCGUAACCACUAG--CUACGCAACGCGACUGUUGCCGG----- --............(((((((((.(((((((((...........))))))))).((((..((((((----------((.......)))))--))))))).......)))))))))----- ( -38.80, z-score = -2.20, R) >dp4.chr4_group2 150858 110 + 1235136 AAAAUACGGCG-CUAU----AUAAGCUGGUAGUGAACCU---CAACUGACAGACUGCGACUGCCAAGCCGCAUACUAGCGUAGCCACUAACUAUACGCAAUGC--UUCGCAUUGAAUUGC .......((((-((..----...)))(((((((.....(---(........)).....))))))).)))(((.....(((((...........)))))(((((--...)))))....))) ( -25.40, z-score = 0.04, R) >droPer1.super_8 227832 110 - 3966273 AAAAUACGGCG-CUAU----AUAAGCUGGUAGUGAACCU---CAACUGACAGACUGCGACUGCCAAGCCGCAUACUAGCGUAGCCACUAACUAUACGCAAUGC--UUCGCAUUGAAUUGC .......((((-((..----...)))(((((((.....(---(........)).....))))))).)))(((.....(((((...........)))))(((((--...)))))....))) ( -25.40, z-score = 0.04, R) >consensus __AAUACGUAGACUUU____AAAAGCUGGCAGUGAACCC___CAGCUGCCAGGCAGCGACUAGCAAGUCGUA____GUCGUAACCACUAG__GUACGCAACGU__CUCUCAUUGAA____ ....((((.................((((((((...........))))))))...((.....))..............))))...................................... (-12.52 = -12.66 + 0.14)


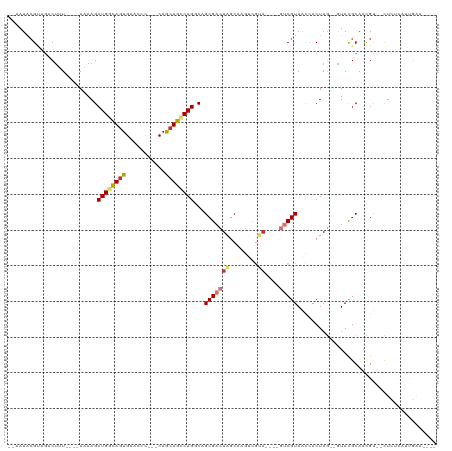
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:07 2011