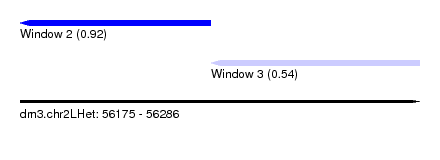
| Sequence ID | dm3.chr2LHet |
|---|---|
| Location | 56,175 – 56,286 |
| Length | 111 |
| Max. P | 0.924536 |

| Location | 56,175 – 56,228 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 71.35 |
| Shannon entropy | 0.51233 |
| G+C content | 0.62582 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -13.18 |
| Energy contribution | -14.46 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
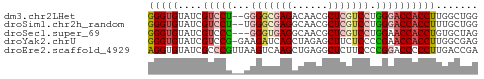
>dm3.chr2LHet 56175 53 - 368872 GGGUGUAUCGUCCU--GGGGCGAGACAACGCUCGUCCUGGGACCACCUUGGCUGG (((((....((((.--.(((((((......)))))))..)))))))))....... ( -27.90, z-score = -3.32, R) >droSim1.chr2h_random 173701 53 + 3178526 GGGUGUAUCGUCCU--UGGGCGAGGCAACGCUCGUCCUGGGACCACCUUUGCUGG (((((....(((((--.(((((((......))))))).))))))))))....... ( -24.80, z-score = -2.67, R) >droSec1.super_69 14338 52 - 174750 GGGUGUAUCGUCCC---GGGUGAGGCAACGCUCGUCCUGGAACCACCUGUGCUAG (((((....((.((---(((((((......)))).))))).)))))))....... ( -18.10, z-score = -0.63, R) >droYak2.chrU 8517614 54 + 28119190 GGGUGUAUCGUCCG-GAAGAUCAGCUAGAGCUUCUCCCCGAACCACCUUGGCGAG (((((..(((...(-((((.((.....)).)))))...)))..)))))....... ( -14.60, z-score = -0.09, R) >droEre2.scaffold_4929 23126963 55 - 26641161 AGGUGUAUCGCCCGUUAAGUCAAGCUGAGGCUCUUCCCCGGACCCCCUUGACCGA .((((...))))......((((((..(.((..(......)..))).))))))... ( -11.30, z-score = 0.60, R) >consensus GGGUGUAUCGUCCG__AGGGCGAGCCAACGCUCGUCCUGGGACCACCUUGGCUGG (((((....(((((...(((((((......))))))).))))))))))....... (-13.18 = -14.46 + 1.28)

| Location | 56,228 – 56,286 |
|---|---|
| Length | 58 |
| Sequences | 5 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 54.83 |
| Shannon entropy | 0.76137 |
| G+C content | 0.62376 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -8.72 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.80 |
| Mean z-score | -0.43 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2LHet 56228 58 - 368872 AUAGCUCCUUGACCCGGACGACGAGUCGGCCCGGCCGGGAU-UAGGAUGUUAUGGGGCA----------- ...(((((.((((((((.(((....)))..))))((.....-..))..)))).))))).----------- ( -22.50, z-score = -0.95, R) >droSim1.chr2h_random 174056 58 + 3178526 CUUGUUCCUCGACGUGGACGACGAGUCGGCCCGGCCGGGAC-UAGGAUGUUAUGGGGCA----------- ..((((((..(((((.(((.....)))(((((....))).)-)...)))))..))))))----------- ( -18.80, z-score = 0.43, R) >droSec1.super_69 14845 58 - 174750 CUGGCUCCCUGACGUGGACGACAAGUCGGUCCGGUCGGGAC-UAGGAUGUUAUGGGGCA----------- ...(((((.((((((.(((.....)))(((((.....))))-)...)))))).))))).----------- ( -23.60, z-score = -1.48, R) >droEre2.scaffold_4929 23127018 52 - 26641161 UUUGCUUCACGAC--------CGUGUCGGCUCGAAUGGGGCUUAGGAUGUUAUGGUGGGG---------- ....((((((.(.--------.(..(((((((.....)))))...))..)..).))))))---------- ( -13.00, z-score = -0.15, R) >droAna3.scaffold_13258 556838 66 + 1584923 -UCGAUCCCCGAUCCCCGCUCCGGGUUGGUGCCAAUACG---CACGCCCUCCCAAAGCUUGCGCCCGGCA -.......(((((((.......)))))))((((....((---((.((.........)).))))...)))) ( -18.70, z-score = -0.01, R) >consensus CUUGCUCCCCGACCCGGACGACGAGUCGGCCCGGACGGGAC_UAGGAUGUUAUGGGGCA___________ ...((((((((((.((.....)).)))))(((....)))..............)))))............ ( -8.72 = -8.68 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:04:42 2011