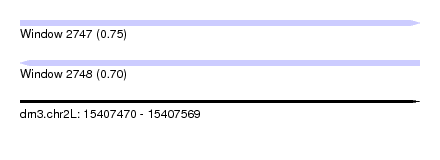
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,407,470 – 15,407,569 |
| Length | 99 |
| Max. P | 0.754364 |

| Location | 15,407,470 – 15,407,569 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.23 |
| Shannon entropy | 0.12323 |
| G+C content | 0.51919 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -30.16 |
| Energy contribution | -30.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
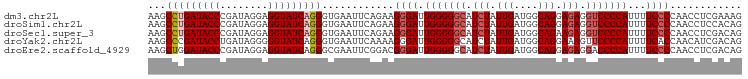
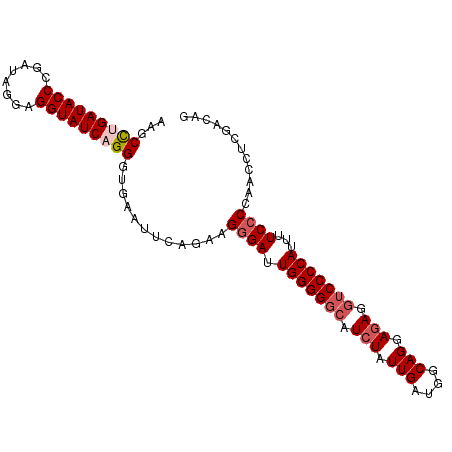
>dm3.chr2L 15407470 99 + 23011544 CUUUCGAGGUUGGGGAAAAUGGGGACCUCUCCUGCCAUCAAUAGAUGCCCCCAAUCCCUUCUGAAUUCACCCUGAUACCUCCUAUCGGGUAUCAGGCUU .....(.((((((((.....((((....))))...((((....)))).)))))))).)............((((((((((......))))))))))... ( -35.90, z-score = -2.18, R) >droSim1.chr2L 15166865 99 + 22036055 CUGUGGAGGUUGGGGAAAAUGGGGACCUCUCCUGCCAUCAAUAGAUGCCCCCAAUCCCUUCUGAAUUCACCCUGAUACCUCCUAUCGGGUAUCAGGCUU ....((.((((((((.....((((....))))...((((....)))).))))))))))............((((((((((......))))))))))... ( -38.70, z-score = -2.23, R) >droSec1.super_3 1667507 99 + 7220098 CUGUCGAGGUUGGGGAAAAUGGGGACCUCUUCUGCCAUCAAUAGAUGCCCCCAAUCCCUUCUGAAUUCACCCUGAUACCUCCUAUCGGGUAUCAGGCUU .....(((...(((((...(((((.(.(((..((....))..))).).))))).)))))......)))..((((((((((......))))))))))... ( -34.00, z-score = -1.49, R) >droYak2.chr2L 2406302 99 - 22324452 CUGUCGAUGUUGGUGAAAAUGGGGAACUUUCCUGCCAUCAAUAGAUGCCCCCAAUCCCUUUUGAAUUCACCCUGAUACCCCCUAUCAGGUAUCGGGCUU ((((.((((..((.(((...((....)))))))..)))).))))..((((.(((......))).......(((((((.....)))))))....)))).. ( -26.40, z-score = -0.84, R) >droEre2.scaffold_4929 7157232 99 + 26641161 CUGUCGAGGUUGGGGAAAAUGGGGUCCUCUCCUGCCAUCAAUAGAUGCCCCCAAUCCCGUCCGAAUUCGCCCUGAUACCUCCUAUCGGGUAUCCAGCUU ..(.((.((((((((.....((((....))))...((((....)))).)))))))).)).).......((...(((((((......)))))))..)).. ( -32.10, z-score = -1.04, R) >consensus CUGUCGAGGUUGGGGAAAAUGGGGACCUCUCCUGCCAUCAAUAGAUGCCCCCAAUCCCUUCUGAAUUCACCCUGAUACCUCCUAUCGGGUAUCAGGCUU .....(.((((((((.....((((....))))...((((....)))).)))))))).)............((((((((((......))))))))))... (-30.16 = -30.72 + 0.56)

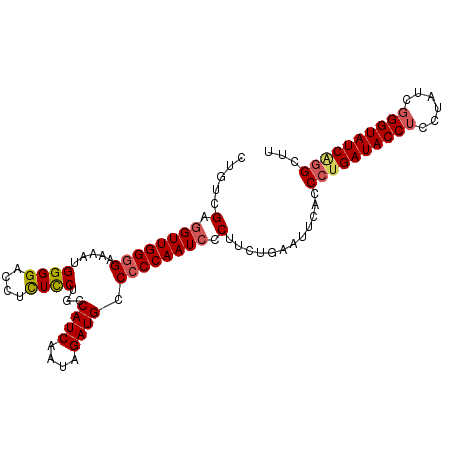
| Location | 15,407,470 – 15,407,569 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.23 |
| Shannon entropy | 0.12323 |
| G+C content | 0.51919 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -31.68 |
| Energy contribution | -32.72 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15407470 99 - 23011544 AAGCCUGAUACCCGAUAGGAGGUAUCAGGGUGAAUUCAGAAGGGAUUGGGGGCAUCUAUUGAUGGCAGGAGAGGUCCCCAUUUUCCCCAACCUCGAAAG ...((((((((((.....).))))))))).....(((((..((((.(((((((.(((.(((....))).))).)))))))...))))....)).))).. ( -38.50, z-score = -2.07, R) >droSim1.chr2L 15166865 99 - 22036055 AAGCCUGAUACCCGAUAGGAGGUAUCAGGGUGAAUUCAGAAGGGAUUGGGGGCAUCUAUUGAUGGCAGGAGAGGUCCCCAUUUUCCCCAACCUCCACAG ...((((((((((.....).)))))))))(((.........((((.(((((((.(((.(((....))).))).)))))))...)))).......))).. ( -39.19, z-score = -2.00, R) >droSec1.super_3 1667507 99 - 7220098 AAGCCUGAUACCCGAUAGGAGGUAUCAGGGUGAAUUCAGAAGGGAUUGGGGGCAUCUAUUGAUGGCAGAAGAGGUCCCCAUUUUCCCCAACCUCGACAG ...((((((((((.....).)))))))))............((((.(((((((.(((.(((....))).))).)))))))...))))............ ( -38.20, z-score = -2.30, R) >droYak2.chr2L 2406302 99 + 22324452 AAGCCCGAUACCUGAUAGGGGGUAUCAGGGUGAAUUCAAAAGGGAUUGGGGGCAUCUAUUGAUGGCAGGAAAGUUCCCCAUUUUCACCAACAUCGACAG ..(((((((((((......))))))).))))..........((((.((((((((((....))))((......))))))))...)).))........... ( -31.10, z-score = -1.17, R) >droEre2.scaffold_4929 7157232 99 - 26641161 AAGCUGGAUACCCGAUAGGAGGUAUCAGGGCGAAUUCGGACGGGAUUGGGGGCAUCUAUUGAUGGCAGGAGAGGACCCCAUUUUCCCCAACCUCGACAG ..(((.(((((((.....).))))))..)))......((..((((.(((((...(((.(((....))).)))...)))))...))))...))....... ( -33.10, z-score = -0.46, R) >consensus AAGCCUGAUACCCGAUAGGAGGUAUCAGGGUGAAUUCAGAAGGGAUUGGGGGCAUCUAUUGAUGGCAGGAGAGGUCCCCAUUUUCCCCAACCUCGACAG ...(((((((((........)))))))))............((((.(((((((.(((.(((....))).))).)))))))...))))............ (-31.68 = -32.72 + 1.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:49 2011