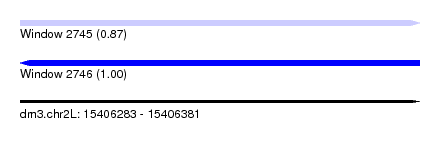
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,406,283 – 15,406,381 |
| Length | 98 |
| Max. P | 0.997658 |

| Location | 15,406,283 – 15,406,381 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.87 |
| Shannon entropy | 0.24348 |
| G+C content | 0.36270 |
| Mean single sequence MFE | -21.81 |
| Consensus MFE | -18.62 |
| Energy contribution | -18.71 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
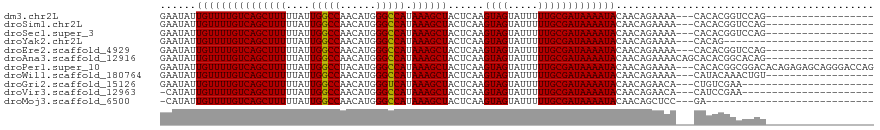
>dm3.chr2L 15406283 98 + 23011544 ------------------CUGGACCGUGUG---UUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC ------------------.(((.(((((.(---(((((.((.(((((((......))))))).))..))).)))..))))).)))(((..((.........))..)))........... ( -22.60, z-score = -1.76, R) >droSim1.chr2L 15165667 98 + 22036055 ------------------CUGGACCGUGUG---UUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC ------------------.(((.(((((.(---(((((.((.(((((((......))))))).))..))).)))..))))).)))(((..((.........))..)))........... ( -22.60, z-score = -1.76, R) >droSec1.super_3 1666311 98 + 7220098 ------------------CUGGACCGUGUG---UUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC ------------------.(((.(((((.(---(((((.((.(((((((......))))))).))..))).)))..))))).)))(((..((.........))..)))........... ( -22.60, z-score = -1.76, R) >droYak2.chr2L 2405086 91 - 22324452 -------------------------CUGUG---UUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC -------------------------...((---((((..((.(((((((......))))))).))....((((((((.(((((......)))))....)))))).))))))))...... ( -21.30, z-score = -2.42, R) >droEre2.scaffold_4929 7156008 98 + 26641161 ------------------CUGGACCGUGUG---UUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC ------------------.(((.(((((.(---(((((.((.(((((((......))))))).))..))).)))..))))).)))(((..((.........))..)))........... ( -22.60, z-score = -1.76, R) >droAna3.scaffold_12916 7711841 101 + 16180835 ------------------CUGUGCCGUGUGCUGUUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC ------------------.((.((((((.(((((((...((.(((((((......))))))).))..)))))))..)))))).))(((..((.........))..)))........... ( -25.70, z-score = -2.44, R) >droPer1.super_10 2027710 116 - 3432795 CUGGUCCCUGCUCUCUGUGUCCGCCGUGUG---UUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUAGGCCAAUAAAAAGCUGACAAAACAAUAUUC ..(((...(((.....)))...)))...((---((((..((.(((((((......))))))).))....((((((((.(((((......)))))....)))))).))))))))...... ( -24.20, z-score = -1.08, R) >droWil1.scaffold_180764 2283898 98 - 3949147 ------------------ACAGUUUGUAUG---UUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC ------------------....(((((...---..(((.((.(((((((......))))))).))..)))(((((((.(((((......)))))....))))))))))))......... ( -21.90, z-score = -1.84, R) >droGri2.scaffold_15126 5606329 94 + 8399593 ----------------------UUCGACAG---UGUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGACCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC ----------------------..((((((---....)))))).......(((.(((..((((.....))))..))).)))....(((..((.........))..)))........... ( -15.50, z-score = -0.43, R) >droVir3.scaffold_12963 12078811 93 + 20206255 ----------------------UUCGGAUG---UGUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUG- ----------------------........---((((.(((((((((((......))))))).........((((((.(((((......)))))....)))))))))).)))).....- ( -21.60, z-score = -1.90, R) >droMoj3.scaffold_6500 20791000 87 + 32352404 ----------------------------UC---GGAGCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUG- ----------------------------..---.....(((((((((((......)))))))...(((..(((((((.(((((......)))))....))))))).))))))).....- ( -19.30, z-score = -1.37, R) >consensus __________________CUGGACCGUGUG___UUUUCUGUUGUAUUUUAUCGCAAAAAUACUACUUGAGUAGCUUUAUGGCCCAUGUUGGCCAAUAAAAAGCUGACAAAACAAUAUUC ......................................(((((((((((......)))))))...(((..(((((((.(((((......)))))....))))))).)))))))...... (-18.62 = -18.71 + 0.09)


| Location | 15,406,283 – 15,406,381 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.87 |
| Shannon entropy | 0.24348 |
| G+C content | 0.36270 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.49 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
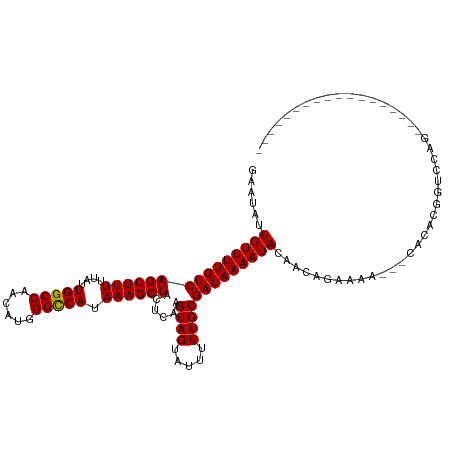
>dm3.chr2L 15406283 98 - 23011544 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAA---CACACGGUCCAG------------------ ......((((((((.((((((....(((((......))))).))))))))....((.(((((((......))))))).))..))))---))..........------------------ ( -24.00, z-score = -2.72, R) >droSim1.chr2L 15165667 98 - 22036055 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAA---CACACGGUCCAG------------------ ......((((((((.((((((....(((((......))))).))))))))....((.(((((((......))))))).))..))))---))..........------------------ ( -24.00, z-score = -2.72, R) >droSec1.super_3 1666311 98 - 7220098 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAA---CACACGGUCCAG------------------ ......((((((((.((((((....(((((......))))).))))))))....((.(((((((......))))))).))..))))---))..........------------------ ( -24.00, z-score = -2.72, R) >droYak2.chr2L 2405086 91 + 22324452 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAA---CACAG------------------------- ......((((((((.((((((....(((((......))))).))))))))....((.(((((((......))))))).))..))))---))...------------------------- ( -24.00, z-score = -3.66, R) >droEre2.scaffold_4929 7156008 98 - 26641161 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAA---CACACGGUCCAG------------------ ......((((((((.((((((....(((((......))))).))))))))....((.(((((((......))))))).))..))))---))..........------------------ ( -24.00, z-score = -2.72, R) >droAna3.scaffold_12916 7711841 101 - 16180835 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAACAGCACACGGCACAG------------------ ......((((((((.((((((....(((((......))))).))))))))....((.(((((((......))))))).))..))))))((.....))....------------------ ( -25.40, z-score = -2.95, R) >droPer1.super_10 2027710 116 + 3432795 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCUACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAA---CACACGGCGGACACAGAGAGCAGGGACCAG .....((((((((((((((((....((((((....)))))).))))))......((.(((((((......))))))).))......---.....)))))).)))).............. ( -28.60, z-score = -2.34, R) >droWil1.scaffold_180764 2283898 98 + 3949147 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAA---CAUACAAACUGU------------------ ......(((((((((((((((....(((((......))))).))))))......((((.....)))))))))))))..((((....---........))))------------------ ( -24.30, z-score = -2.97, R) >droGri2.scaffold_15126 5606329 94 - 8399593 GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGUCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAACA---CUGUCGAA---------------------- ((....((((((((.((((((....(((((......))))).)))))).........(((((((......))))))).))))))))---...))...---------------------- ( -24.40, z-score = -3.02, R) >droVir3.scaffold_12963 12078811 93 - 20206255 -CAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAACA---CAUCCGAA---------------------- -.....((((((((.((((((....(((((......))))).)))))).........(((((((......))))))).))))))))---........---------------------- ( -26.20, z-score = -4.21, R) >droMoj3.scaffold_6500 20791000 87 - 32352404 -CAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGCUCC---GA---------------------------- -.....(((((((((((((((....(((((......))))).))))))......((((.....)))))))))))))..........---..---------------------------- ( -23.70, z-score = -3.08, R) >consensus GAAUAUUGUUUUGUCAGCUUUUUAUUGGCCAACAUGGGCCAUAAAGCUACUCAAGUAGUAUUUUUGCGAUAAAAUACAACAGAAAA___CACACGGUCCAG__________________ ......(((((((((((((((....(((((......))))).))))))......((((.....)))))))))))))........................................... (-23.57 = -23.49 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:47 2011