| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,400,730 – 15,400,837 |
| Length | 107 |
| Max. P | 0.970591 |

| Location | 15,400,730 – 15,400,837 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.89 |
| Shannon entropy | 0.60216 |
| G+C content | 0.34567 |
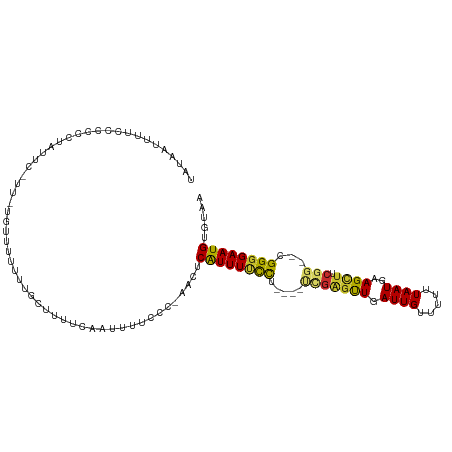
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -9.70 |
| Energy contribution | -9.36 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
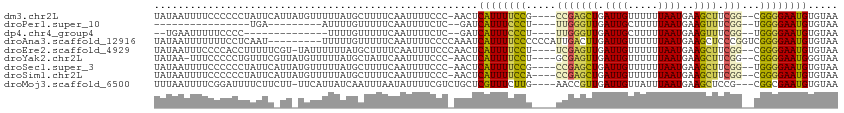
>dm3.chr2L 15400730 107 + 23011544 UAUAAUUUUCCCCCCUAUUCAUUAUGUUUUUAUGCUUUUCAAUUUUCCC-AACUCAUUUUCCG----CCGAGCUGAUUGUUUUUUAAUGAAGCUUCGG--CGGGGAAUGUGUAA .................................................-.((.((((..(((----(((((((.((((.....))))..))).))))--)))..)))).)).. ( -27.80, z-score = -3.70, R) >droPer1.super_10 2021916 81 - 3432795 ----------------UGA---------AUUUUGUUUUUCAAUUUUCUC--GAUCAUUUCCCU----UUGGGUUGAUUGCUUUUUAAUGAAGUUUCGG--UGGGGAAUGUGUAA ----------------.((---------(..(((.....)))..)))..--...((((((((.----(((((......(((((.....))))))))))--.))))))))..... ( -16.40, z-score = -0.95, R) >dp4.chr4_group4 4185472 90 - 6586962 --UGAAUUUUUCCCC--------------UUUUGUUUUUCAAUUUUCUC--GAUCAUUUCCCU----UUGGGUUGAUUGCUUUUUAAUGAAGUUUCGG--UGGGGAAUGUGUAA --.......((((((--------------..(((..(((((.......(--(((((...(((.----..))).))))))........)))))...)))--.))))))....... ( -20.86, z-score = -1.71, R) >droAna3.scaffold_12916 7706361 105 + 16180835 UAUAAUUUUUUUCCUCAAU---------UUUUUGUUUUUCAAUUUUCCCCAAAUCAUUUUCCCCCCAUUGACUUGAUUGUUUUUUAAUGAAGCUCCCGGUCGGGGAAUGUGUAA ...................---------..........................(((.((((((((...(((((.((((.....)))).))).))..))..)))))).)))... ( -16.60, z-score = -0.44, R) >droEre2.scaffold_4929 7150214 107 + 26641161 UAUAAUUUCCCCACCUUUUUCGU-UAUUUUUUAUGCUUUUCAAUUUUCCCAACUCAUUUUCCU----UCGAGUUGAUUGUUUUUUAAUGAAGCUUCGG--CGGGGAAUGUGUAA ((((..((((((......(((((-((.......((.....)).......((((((........----..)))))).........)))))))((....)--)))))))..)))). ( -25.10, z-score = -3.46, R) >droYak2.chr2L 2399351 106 - 22324452 UAUAA-UUUCCCCCUGUUUCGUUAUGUUUUUAUGCUAUUCAAUUUUCCC-AACUCAUUUUCCU----GCGAGUUGAUUGUUUUUUAAUGAAGCUUCGG--CGGGGAAUGGGUAA (((.(-((((((((.(((((((((.((......)).............(-(((((........----..)))))).........)))))))))...))--.)))))))..))). ( -25.60, z-score = -1.96, R) >droSec1.super_3 1660737 107 + 7220098 UAUAAUUUUCCCCCCUAUUCAUUAUGUUUUUAUGCUUUUCAAUUUUCCC-AACUCAUUUUCCG----CCGAGCUGAUUGUUUUUUAAUGAAGCUUCGG--UGGGGAAUGUGUAA .................................................-.((.((((..(((----(((((((.((((.....))))..))).))))--)))..)))).)).. ( -25.90, z-score = -3.22, R) >droSim1.chr2L 15160034 107 + 22036055 UAUAAUUUUCCCCCCUAUUCAUUAUGUUUUUAUGCUUUUCAAUUUUCCC-AACUCAUUUUCCA----CCGAGCUGAUUGUUUUUUAAUGAAGCUUCGG--CGGGGAAUGUGUAA .................................................-.((.((((..((.----(((((((.((((.....))))..))).))))--.))..)))).)).. ( -22.40, z-score = -2.09, R) >droMoj3.scaffold_6500 20784622 106 + 32352404 UUUAAUUUUCGGAUUUUCUUCUU-UUCAUUAUCAAUUUAAUAUUUUCGUCUGCUCGUUUCUUG----AACCGUUGAUUGUUAUUUAAUGAAGCUCCG---CGGCGAAUGUGUAA ..........(((......))).-...............((((.((((.((((..((((((((----((.((.....))...))))).)))))...)---))))))).)))).. ( -14.90, z-score = 0.25, R) >consensus UAUAAUUUUCCCCCCUAUUC_UU_UGUUUUUUUGCUUUUCAAUUUUCCC_AACUCAUUUUCCU____UCGAGUUGAUUGUUUUUUAAUGAAGCUUCGG__CGGGGAAUGUGUAA ......................................................((((((((.....(((((((.((((.....))))..)))).)))...))))))))..... ( -9.70 = -9.36 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:45 2011