
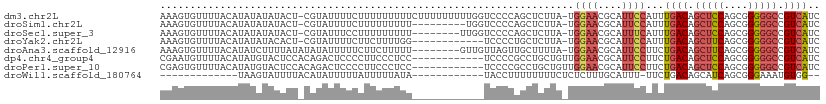
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,397,186 – 15,397,294 |
| Length | 108 |
| Max. P | 0.955073 |

| Location | 15,397,186 – 15,397,294 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 69.98 |
| Shannon entropy | 0.57018 |
| G+C content | 0.43006 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.26 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.955073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
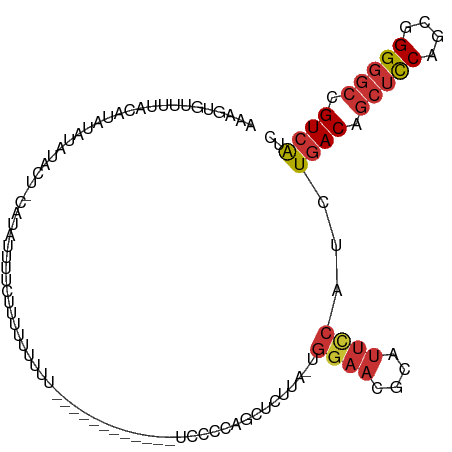
>dm3.chr2L 15397186 108 - 23011544 AAAGUGUUUUACAUAUAUAUACU-CGUAUUUUCUUUUUUUUUCUUUUUUUUUGGUCCCCAGCUCUUA-UGGAACGCAUUCCAUUUGACAGCUCCAGCGGGGGCCGUCAUC ..(((((...........)))))-............................(((((((.(.((..(-(((((....))))))..))).((....)))))))))...... ( -25.20, z-score = -2.90, R) >droSim1.chr2L 15156579 99 - 22036055 AAAGUGUUUUACAUAUAUAUACU-CGUAUUUUCUUUUUUUUU---------UGGUCCCCAGCUCUUA-UGGAACGCAUUCCAUUUGACAGCUCCAGCGGGGGCCGUCAUC ..(((((...........)))))-..................---------.(((((((.(.((..(-(((((....))))))..))).((....)))))))))...... ( -25.20, z-score = -2.79, R) >droSec1.super_3 1657267 100 - 7220098 AAAGUGUUUUACAUAUAUAUACU-CGUAUUUCCUUUUUUUUU--------UUGGUCCCCAGCUCUUA-UGGAACGCAUUUCAUUUGACAGCUCCAGCGGGGGCCGUCAUC ..(((((...........)))))-..................--------..(((((((.(.((..(-(((((....))))))..))).((....)))))))))...... ( -22.30, z-score = -1.71, R) >droYak2.chr2L 2395898 96 + 22324452 AAAGUGUUUUACAUAUAUACACU-CGUAUUUUCUUCUUUUGG------------UCCCCUGCUCUUA-UGGAACGCAUUCCAUUUGACAGCUUCAGCGGGGGCCGUCAUC ..(((((...........)))))-................((------------(((((((.((..(-(((((....))))))..))))((....)))))))))...... ( -27.20, z-score = -3.39, R) >droAna3.scaffold_12916 7702776 101 - 16180835 AAAGUGUUUUACAUAUCUUUUAUAUAUAUUUUUCUUCUUUUU--------GUUGUUAGUUGCUUUUA-UGGAACGCAUUCCUUCUGACAGCUUCAGCGGGGGCCGUCAUC ...((((((((.(((.....)))...................--------...((.....)).....-))))))))........((((.(((((....))))).)))).. ( -14.50, z-score = 0.42, R) >dp4.chr4_group4 4183185 98 + 6586962 CGAAUGUUUUACAUAUGUACUCCACAGACUCCCCUUCCCUCC------------UCCCCGCCUGCUGUUGGAACGCAUUCCUUCUGACAGCUCCAGCGGGGGCCGUCAUC .((.(((..(((....)))....)))................------------(((((((..(((((..(((........)))..)))))....)))))))...))... ( -26.70, z-score = -1.73, R) >droPer1.super_10 2019396 98 + 3432795 CGAGUGUUUUACAUAUGUACUCCACAGACUCCCCUUCCCUCC------------UCCCCGCCUGCUGUUGGAACGCAUUCCUUCUGACAGCUCCAGCGGGGGCCGUCAUC .(((((...(((....)))...))).................------------(((((((..(((((..(((........)))..)))))....)))))))...))... ( -29.70, z-score = -2.18, R) >droWil1.scaffold_180764 1671922 82 - 3949147 -------------UAAGUAUUUUACAUAUUUUUAUUUUUAUA------------UACCUUUUUUUUCUCUCUUUGCAUUU-UUCUGACAGCAUCAGCGGGAAAUGUGG-- -------------.............................------------..................(..(((((-((((((.....))))..)))))))..)-- ( -8.20, z-score = -0.16, R) >consensus AAAGUGUUUUACAUAUAUAUACU_CAUAUUUUCUUUUUUUUU____________UCCCCAGCUCUUA_UGGAACGCAUUCCAUCUGACAGCUCCAGCGGGGGCCGUCAUC .....................................................................((((....))))...((((.(((((....))))).)))).. (-10.70 = -11.26 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:44 2011