| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,391,110 – 15,391,201 |
| Length | 91 |
| Max. P | 0.995929 |

| Location | 15,391,110 – 15,391,201 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 70.47 |
| Shannon entropy | 0.59689 |
| G+C content | 0.44941 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -12.30 |
| Energy contribution | -12.12 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.995929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15391110 91 - 23011544 UCGAUGAGCGAGUGUGUUGACGCACUUGCCUCCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCAGAGU-----GGAGAGCUGGCGGGGCAC------- .......(((((((((....)))))))))(((((((((....)))))..(((.((((((....)))))).)))-----)))).(((.....)))..------- ( -30.70, z-score = -3.46, R) >droSim1.chr2L 15150513 91 - 22036055 UCGAUGAGCGAGUGUGUUGACGCACUUGCCUCCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCAGAGU-----GGGGAGCUGGCGGGGCAC------- .......(((((((((....)))))))))(((((((((....)))))..(((.((((((....)))))).)))-----)))).(((.....)))..------- ( -29.80, z-score = -2.90, R) >droSec1.super_3 1651231 91 - 7220098 UCGAUGAGCGAGUGUGUUGACGCACUUGCCUCCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCAGAGU-----GGGGAGCUGGCGGGGCCC------- .......(((((((((....)))))))))(((((((((....))))...(((.((((((....)))))).)))-----.)))))..(((...))).------- ( -29.90, z-score = -2.58, R) >droYak2.chr2L 2389744 103 + 22324452 UCGAUGAGCGAGUGUGUUGACGCACUUGCCUCCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCAGAGUGGGGAGGGGAGCUGGUGGGGCACUGGAGUG ((.((.((((((((((....))))))).((((((((((....))))...(((.((((((....)))))).))).))))))...))).)).)).(((....))) ( -34.90, z-score = -3.49, R) >droEre2.scaffold_4929 7140616 97 - 26641161 CCGAUGAGCGUGUGUGUUGACGCACUUGCCUCCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCAGGGA-----GGGGAGCUGGCGGGGCACUGGCAC- (((.(.(((((((((....))))))((.((((((((((....)))).......((((((....))))))))))-----)).))))).)))).((....))..- ( -34.00, z-score = -2.75, R) >dp4.chr4_group4 4177893 85 + 6586962 --------UUUCUG-ACUGACGCACUUGCCUUCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCUGAAAGAG--GGAGGUGCAGAGGGGCAG------- --------...(((-.((...((((((.((((((((((....))))).......(((((....))))).....)))--))))))))...))..)))------- ( -23.90, z-score = -2.45, R) >droPer1.super_10 2014121 85 + 3432795 --------UUUCUG-ACUGACGCACUUGCCUUCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCUGAAAGAG--GGAGGUGCAGCGGGGCAC------- --------......-.(((..((((((.((((((((((....))))).......(((((....))))).....)))--))))))))..))).....------- ( -24.30, z-score = -2.60, R) >droWil1.scaffold_180764 1689771 90 - 3949147 -------------UUGCUCAGGCGGCCUUUUUCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCUAAAAGAAAACCCCUGCAAGUAGAAGGCAAAGUAA -------------(((((((((.((.(((((.(((((..((((((.......))))))....)))))..)))))....)))))).........)))))..... ( -18.10, z-score = -0.81, R) >droMoj3.scaffold_6500 11578917 81 + 32352404 ----UGGAGCUGCCUGUUGACGCA-AUGCCUCCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCUAAGC-----GCGGCAGCGGCUG------------ ----...((((((.(((((.(((.-........(((((....))))).......(((((....)))))...))-----)))))))))))).------------ ( -25.80, z-score = -2.22, R) >consensus _CGAUGAGCGAGUGUGUUGACGCACUUGCCUCCUAAUGAUAACAUUAAAAUUGUGUUAAGAAAUUAGCAGAGA_____GGGGAGCUGGCGGGGCAC_______ .............((((....)))).((((((.(((((....))))).......(((((....))))).....................))))))........ (-12.30 = -12.12 + -0.18)

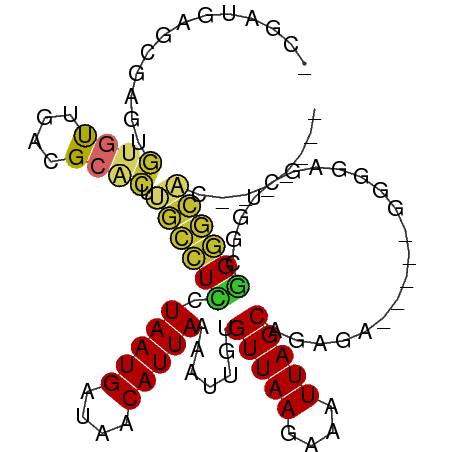
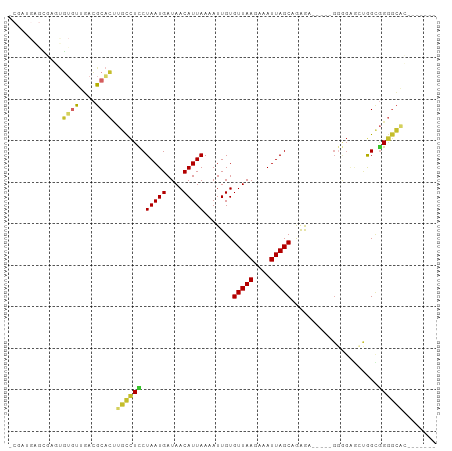
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:41 2011