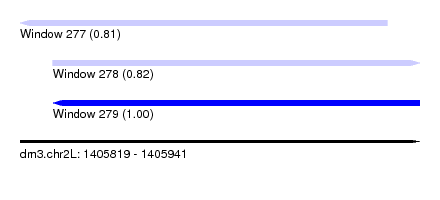
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,405,819 – 1,405,941 |
| Length | 122 |
| Max. P | 0.995545 |

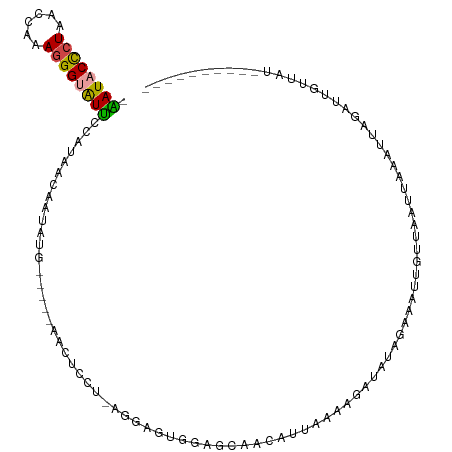
| Location | 1,405,819 – 1,405,931 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 53.08 |
| Shannon entropy | 0.84907 |
| G+C content | 0.32602 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -7.54 |
| Energy contribution | -7.32 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1405819 112 - 23011544 -GAUACUCUAAGCGAAGGGUAUUUUAAAACCUUAUGAGCUCAGCUCCU-AAGAGAGGAACAACAUUAAAAGAUAUAGAAAUUGUUAAUUAAAUUAUGUUCUUAUGGAUCAUUAC -((((((((......)))))))).......((((.((((...)))).)-)))..(((((((...((((..((((.......))))..))))....)))))))............ ( -19.10, z-score = -0.48, R) >droSim1.chr2L 1379072 94 - 22036055 -AAUACCCUAACCAAAGGGUAUUCAUUAACAAUAUG-----AUCGCCU-AGGAG-------------AAAGAUAUAGAAAUUGUUAAUUAAAUUAAAUUGUUAUGGAUCAUUGC -..((((((......))))))(((((.((((((.((-----(((.(..-..).)-------------)..((((.......)))).......))).)))))))))))....... ( -15.00, z-score = -0.52, R) >droSec1.super_14 1353520 107 - 2068291 -AAUACCCUAACCAAAGGGUAUUCAUUAACAAUAUG-----AUCUCCU-AGGAGUGGAACAACAUUAAAAGAUAUAGAAGUUGUUAAUUCAAUUAAAUUGUUAUGGAUCAUUGU -..((((((......))))))(((((.((((((.((-----(((((..-..)))...((((((................))))))......)))).)))))))))))....... ( -21.69, z-score = -1.26, R) >droYak2.chr2L 1380250 96 - 22324452 -GAUACCCUAAACGCAGGGUAUCUCAAAACAUUCAG-----AGCUCUUGGGGAGUGAAGCAAUAUUACAAGAUAUAGCAAUAAUGGGAUAG--UGGAUCGUUAA---------- -((((((((......))))))))....((((((((.-----.((((.....))))...((.(((((....))))).)).............--))))).)))..---------- ( -23.20, z-score = -1.84, R) >dp4.chr4_group3 9924161 102 - 11692001 GAAUACCCUAUCGAAAGGGGGUUGCAUUGAAAUUCGAUAUACAUUUUAUAGGGUUAUAGAAUCAUAAUUUGAUGCUGCGAUUGCUUGUAAUUGCAGAUUUGU------------ ....(((((((.((((....((...((((.....))))..)).)))))))))))......((((.....)))).(((((((((....)))))))))......------------ ( -23.20, z-score = -1.20, R) >droPer1.super_8 1131488 101 - 3966273 GAAUACCCUAUCGAAA-GGGGUUGCAUUGAAAUUCGAUAUACAUUUUAUAGGCAUAUAGAAUCAUAAUUUGAUGCUGCGAUUGCUUGUAAUUGGAGAUUUGU------------ (...(((((.......-)))))..)((((.....))))...((..(((((((((..(((.((((.....)))).)))....))))))))).)).........------------ ( -20.20, z-score = -0.44, R) >consensus _AAUACCCUAACCAAAGGGUAUUCCAUAACAAUAUG_____AACUCCU_AGGAGUGGAGCAACAUUAAAAGAUAUAGAAAUUGUUAAUUAAAUUAGAUUGUUAU__________ .((((((((......))))))))........................................................................................... ( -7.54 = -7.32 + -0.22)

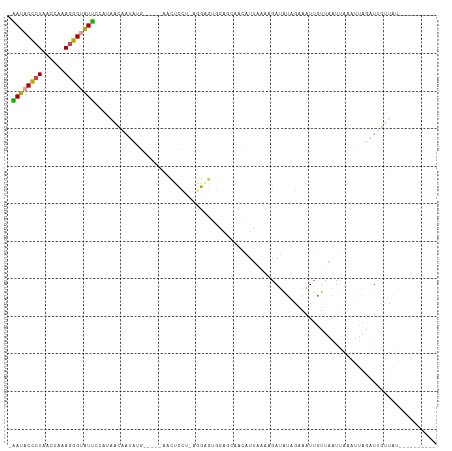
| Location | 1,405,829 – 1,405,941 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 55.61 |
| Shannon entropy | 0.79049 |
| G+C content | 0.33409 |
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -5.17 |
| Energy contribution | -6.12 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1405829 112 + 23011544 AUAAGAACAUAAUUUAAUUAACAAUUUCUAUAUCUUUUAAUGUUGUUCCUCUCUUAGGAGCUG-AGCUCAUAAGGUUUUAAAAUACCCUUCGCUUAGAGUAUCCACUUAGCGA- ...(((((...........((((((................)))))).....((((.((((..-.)))).)))))))))..........(((((..((((....)))))))))- ( -16.99, z-score = -0.04, R) >droSim1.chr2L 1379082 94 + 22036055 AUAACAAUUUAAUUUAAUUAACAAUUUCUAUAUCUUU-------------CUCCUAGG------CGAUCAUAUUGUUAAUGAAUACCCUUUGGUUAGGGUAUUCACUUAGCGA- ................(((((((((.......(((..-------------.....)))------.......)))))))))(((((((((......))))))))).........- ( -19.64, z-score = -2.91, R) >droSec1.super_14 1353530 107 + 2068291 AUAACAAUUUAAUUGAAUUAACAACUUCUAUAUCUUUUAAUGUUGUUCCACUCCUAGG------AGAUCAUAUUGUUAAUGAAUACCCUUUGGUUAGGGUAUUCACUUAGCGA- .(((((((.....(((...((((((................))))))...(((....)------)).))).))))))).((((((((((......))))))))))........- ( -26.49, z-score = -3.40, R) >droYak2.chr2L 1380260 96 + 22324452 ------------ACUAUCCCAUUAUUGCUAUAUCUUGUAAUAUUGCUUCACUCCCCAA----G-AGCUCUGAAUGUUUUGAGAUACCCUGCGUUUAGGGUAUCCACUUAGCGA- ------------.......((.((((((........)))))).)).(((((((.....----)-))...))))((((.((.(((((((((....)))))))))))...)))).- ( -21.20, z-score = -1.95, R) >dp4.chr4_group3 9924171 102 + 11692001 ------------AAUUACAAGCAAUCGCAGCAUCAAAUUAUGAUUCUAUAACCCUAUAAAAUGUAUAUCGAAUUUCAAUGCAACCCCCUUUCGAUAGGGUAUUCCAUUCACCAG ------------...((((.((....))((.((((.....)))).))..............))))...........((((.((..((((......))))..)).))))...... ( -12.20, z-score = -0.68, R) >droPer1.super_8 1131498 101 + 3966273 ------------AAUUACAAGCAAUCGCAGCAUCAAAUUAUGAUUCUAUAUGCCUAUAAAAUGUAUAUCGAAUUUCAAUGCAACCCC-UUUCGAUAGGGUAUUCCAUUCACCAG ------------........((....)).((((........(((((.((((((.........)))))).)))))...))))...(((-(......))))............... ( -14.90, z-score = -1.03, R) >consensus ____________AUUAACUAACAAUUGCUAUAUCUUUUAAUGUUGCUACACUCCUAGA____G_AGAUCAUAUUGUUAUGAAAUACCCUUUGGUUAGGGUAUUCACUUAGCGA_ .................................................................................((((((((......))))))))........... ( -5.17 = -6.12 + 0.95)

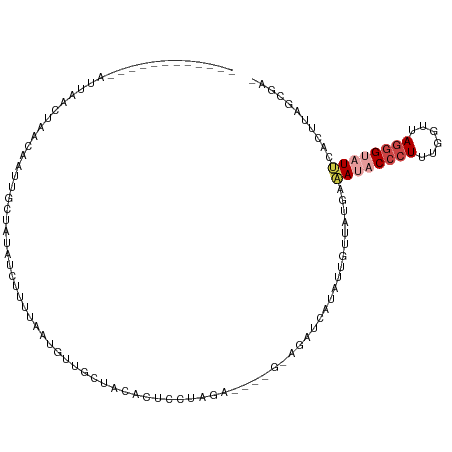
| Location | 1,405,829 – 1,405,941 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 55.61 |
| Shannon entropy | 0.79049 |
| G+C content | 0.33409 |
| Mean single sequence MFE | -22.25 |
| Consensus MFE | -10.19 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
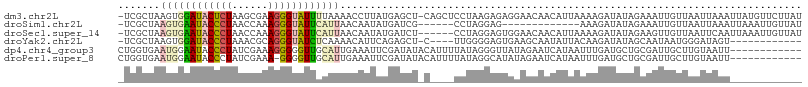
>dm3.chr2L 1405829 112 - 23011544 -UCGCUAAGUGGAUACUCUAAGCGAAGGGUAUUUUAAAACCUUAUGAGCU-CAGCUCCUAAGAGAGGAACAACAUUAAAAGAUAUAGAAAUUGUUAAUUAAAUUAUGUUCUUAU -(((((.((.(....).)).)))))...............((((.((((.-..)))).))))..(((((((...((((..((((.......))))..))))....))))))).. ( -20.90, z-score = -0.59, R) >droSim1.chr2L 1379082 94 - 22036055 -UCGCUAAGUGAAUACCCUAACCAAAGGGUAUUCAUUAACAAUAUGAUCG------CCUAGGAG-------------AAAGAUAUAGAAAUUGUUAAUUAAAUUAAAUUGUUAU -......((((((((((((......))))))))))))((((((.(((((.------(....).)-------------)..((((.......)))).......))).)))))).. ( -20.10, z-score = -2.94, R) >droSec1.super_14 1353530 107 - 2068291 -UCGCUAAGUGAAUACCCUAACCAAAGGGUAUUCAUUAACAAUAUGAUCU------CCUAGGAGUGGAACAACAUUAAAAGAUAUAGAAGUUGUUAAUUCAAUUAAAUUGUUAU -......((((((((((((......))))))))))))((((((.((((((------(....)))...((((((................))))))......)))).)))))).. ( -26.79, z-score = -3.62, R) >droYak2.chr2L 1380260 96 - 22324452 -UCGCUAAGUGGAUACCCUAAACGCAGGGUAUCUCAAAACAUUCAGAGCU-C----UUGGGGAGUGAAGCAAUAUUACAAGAUAUAGCAAUAAUGGGAUAGU------------ -..((((..((((((((((......)))))))).))...((((....(((-(----.....))))...((.(((((....))))).))...))))...))))------------ ( -23.10, z-score = -1.56, R) >dp4.chr4_group3 9924171 102 - 11692001 CUGGUGAAUGGAAUACCCUAUCGAAAGGGGGUUGCAUUGAAAUUCGAUAUACAUUUUAUAGGGUUAUAGAAUCAUAAUUUGAUGCUGCGAUUGCUUGUAAUU------------ ....(.((((.((..((((......))))..)).)))).)...............(((((((((..(((.((((.....)))).)))..))..)))))))..------------ ( -20.30, z-score = -0.24, R) >droPer1.super_8 1131498 101 - 3966273 CUGGUGAAUGGAAUACCCUAUCGAAA-GGGGUUGCAUUGAAAUUCGAUAUACAUUUUAUAGGCAUAUAGAAUCAUAAUUUGAUGCUGCGAUUGCUUGUAAUU------------ ....(.((((.(((.((((......)-)))))).)))).)...............(((((((((..(((.((((.....)))).)))....)))))))))..------------ ( -22.30, z-score = -1.04, R) >consensus _UCGCUAAGUGAAUACCCUAACCAAAGGGUAUUCCAUAACAAUAUGAUCU_C____CCUAGGAGUGGAGCAACAUUAAAAGAUAUAGAAAUUGUUAAUUAAA____________ .......((((((((((((......))))))))))))............................................................................. (-10.19 = -10.75 + 0.56)
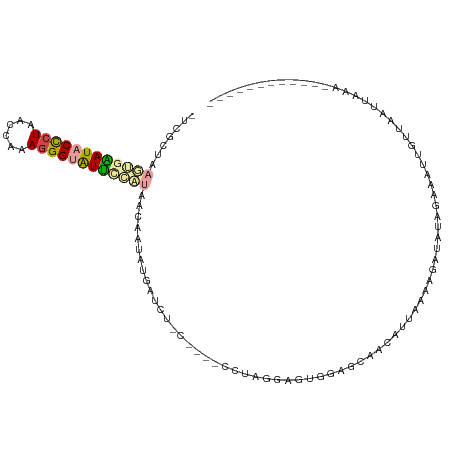
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:31 2011