| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,378,168 – 15,378,284 |
| Length | 116 |
| Max. P | 0.996577 |

| Location | 15,378,168 – 15,378,284 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Shannon entropy | 0.44517 |
| G+C content | 0.32484 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -15.24 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
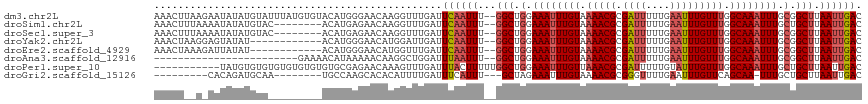
>dm3.chr2L 15378168 116 + 23011544 AAACUUAAGAAUAUAUGUAUUUAUGUGUACAUGGGAACAAGGUUUGAUUCAAUUU--GGCUGGAAAUUUGUAAAACGCGAUUUUUGAAUUUGUUUGGCAAAUUUGCGGCUUAAUUGAC ((((((.......(((((((......)))))))......))))))...((((((.--(((((.((((((((.(((((.((((....))))))))).)))))))).))))).)))))). ( -31.62, z-score = -4.29, R) >droSim1.chr2L 15136779 108 + 22036055 AAACUUUAAAAUAUAUGUAC--------ACAUGAGAACAAGGUUUGAUUCAAUUU--GGCUGGAAAUUUGUAAAACGCGAUUUUUGAAUUUGUUUGGCAAAUUUGCUGCUUAAUUGAC (((((((......((((...--------.)))).....)))))))...((((((.--(((.(.((((((((.(((((.((((....))))))))).)))))))).).))).)))))). ( -23.70, z-score = -2.24, R) >droSec1.super_3 1638309 108 + 7220098 AAACUUUAAAAUAUAUGUAC--------ACAUGAGAACAAGGUUUGAUUCAAUUU--GGCUGGAAAUUUGUAAAACGCGAUUUUUGAAUUUGUUUGGCAAAUUUGCGGCUUAAUUGAC (((((((......((((...--------.)))).....)))))))...((((((.--(((((.((((((((.(((((.((((....))))))))).)))))))).))))).)))))). ( -27.60, z-score = -3.59, R) >droYak2.chr2L 2376006 104 - 22324452 AAACUAAGGAGUAUAU------------ACAUGGGAACAUGGAUUGAUUCAAUUU--GGCUGGAAAUUUGUAAAACGCGAUUUUUGAAUUUGUUUGGCAAAUUUGCGGCUUAAUUGAC ................------------.((((....)))).......((((((.--(((((.((((((((.(((((.((((....))))))))).)))))))).))))).)))))). ( -29.20, z-score = -4.83, R) >droEre2.scaffold_4929 7127595 104 + 26641161 AAACUAAAGAUUAUAU------------ACAUGGGAACAUGGUUUGAUUCAAUUU--GGCUGGAAAUUUGUAAAACGCGAUUUUUGAAUUUGUUUGGCAAAUUUGCGGCUUAAUUGAC ................------------.((((....)))).......((((((.--(((((.((((((((.(((((.((((....))))))))).)))))))).))))).)))))). ( -29.20, z-score = -4.86, R) >droAna3.scaffold_12916 7684772 92 + 16180835 ------------------------GAAAACAUAAAAACAAGGCUGGAUUUAAUUU--GGCUGGAAAUUUGUAAAACGCGAUUUUUGAAUUUGUUUGGCAAAUUUGCGGCUUAAUUGAC ------------------------........................((((((.--(((((.((((((((.(((((.((((....))))))))).)))))))).))))).)))))). ( -21.60, z-score = -2.85, R) >droPer1.super_10 2000342 107 - 3432795 -----------UAUGUGUGUGUGUGUGUGUGCGAGAACAAAGUUUGAUUUACUUUUUGGCUGGAAAUUUGUUAAACGCGAUUUUUGUAUUUGUUUGGCAAAUUUGCUGCUUAAUUGAC -----------..(((...((..(....)..))...)))............(..((.(((.(.((((((((((((((.(((......))))))))))))))))).).))).))..).. ( -21.20, z-score = -0.26, R) >droGri2.scaffold_15126 5578067 97 + 8399593 ---------CACAGAUGCAA--------UGCCAAGCACACAUUUUGAUUUCAUUU---GCUAGAAAUUUGUAAAACGCGGGUUUUGAAUUUGUUCAGCAA-UUUGCUGCUUAAUUGAC ---------.((((((.(((--------.(((..((.........((((((....---....))))))........)).))).))).)))))).((((..-...)))).......... ( -18.13, z-score = 0.63, R) >consensus AAACU__A_AAUAUAUGUA_________ACAUGAGAACAAGGUUUGAUUCAAUUU__GGCUGGAAAUUUGUAAAACGCGAUUUUUGAAUUUGUUUGGCAAAUUUGCGGCUUAAUUGAC ................................................((((((...(((.(.((((((((.(((((.((((....))))))))).)))))))).).))).)))))). (-15.24 = -15.90 + 0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:36 2011