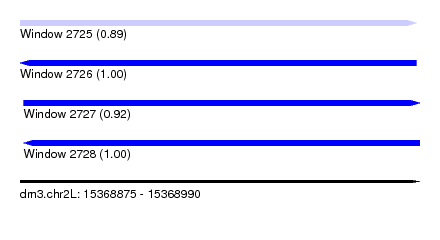
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,368,875 – 15,368,990 |
| Length | 115 |
| Max. P | 0.999778 |

| Location | 15,368,875 – 15,368,989 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Shannon entropy | 0.52123 |
| G+C content | 0.54101 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -19.66 |
| Energy contribution | -19.99 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15368875 114 + 23011544 GCUCGAGACUU-GAGAGGU--AGGUUCACCUGUUUAUGCCAGAUUACUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGAAGUCUUGGAGUCGCAACUCC ....(((((((-((.((((--(((....)))...((((((((....))))))))...........))))((....))..(((((....))))))).)))))))(((((....))))) ( -37.80, z-score = -1.75, R) >droSim1.chr2L 15127294 114 + 22036055 GCUCGAGACUU-GAGAGGU--AGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGAAGUCUUGGAGUCGCAACUCC ....(((((((-((.((((--(((....)))...((((((((....))))))))...........))))((....))..(((((....))))))).)))))))(((((....))))) ( -38.20, z-score = -1.78, R) >droSec1.super_3 1629178 114 + 7220098 GCUCGAGACUU-GAGAGGU--AGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACACAGUCGAAGUCUUGGAGUCGCAACUCC ....(((((((-((.((((--(((....)))...((((((((....))))))))...........))))((((((....))))))........)).)))))))(((((....))))) ( -37.30, z-score = -1.90, R) >droYak2.chr2L 2366674 114 - 22324452 GCUUGAGACUU-GAGAGGU--AGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUAGAAGUCUUGGAGUCGCAACUCC ..(((.(((((-.....((--((((.........((((((((....)))))))).(((....))))))))).(((((((((((((....)).))))))).))))))))).))).... ( -38.60, z-score = -2.09, R) >droEre2.scaffold_4929 7118505 114 + 26641161 ACUUGAGACUU-GAGAGGU--AGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCAUCUGCGCAAGCUUUUGCGCCCACGCAGUGGAAGUCUUGGAGUCGCAACUCC ....(((((((-.((((((--(((....)))...((((((((....))))))))........))).)))((((((....))))))((((...)))))))))))(((((....))))) ( -37.60, z-score = -1.67, R) >droAna3.scaffold_12916 7675661 95 + 16180835 GGUGGAGUCUU-CGGUGG------UUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGCAGUCC--------------- ((((((.((..-..).).------)))))).....(((((((....)))))))((((((..........((....))....(((....))))))))).....--------------- ( -34.30, z-score = -2.25, R) >dp4.chr4_group4 4157381 102 - 6586962 GUCGGAGUCUU-CGGAGG------UUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCUUUUGCGCCCACGCAGCCGCAGCCUCAGCCUC-------- ....(((.((.-((((((------....)))...((((((((....)))))))).............(((((..(((......)))..)))))))).)).)))......-------- ( -34.70, z-score = -1.24, R) >droPer1.super_10 1993426 108 - 3432795 GUCGGAGUCUU-CGGAGG------UUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCUUUUGCGCCCACGCAGCCGAAGCCGCAGCCGCAGCCUC-- (.(((.(.(((-((((((------....)))...((((((((....)))))))).............(((((..(((......)))..))))))))))).)....))))......-- ( -38.80, z-score = -1.28, R) >droWil1.scaffold_180764 2214380 96 - 3949147 AGUUGAGUUGA-GGUAAGGCGAGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUUGG-------------------- ..........(-(((...((.(((....)))....(((((((....)))))))))(((....)))))))((....))..(((((....)))))....-------------------- ( -27.70, z-score = -0.11, R) >droMoj3.scaffold_6500 11603764 106 - 32352404 CCCUGGGUCUUCGGUUAGACUUGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCA--UGCGCCCACGCAGCCUCACA--------GUUGCCCC- ..(..(((((......)))))..)..((.((((.((((((((....)))))))).............(((((..(((.--...)))..)))))....)))--------).))....- ( -35.30, z-score = -1.52, R) >droVir3.scaffold_12963 8171817 113 - 20206255 CCCUCAGUCUU-GGUUAGACUUGGUUCACCUGUUUAUGCCAGAUUCCAGGCAUGCGAUUAAAAUCACCUGCGCAGGCA--UGCGCCCACGCAGCCUCGCAGCCUCGGAGUUGCCUG- ....(((...(-((((((((..((....)).))))).))))((((((((((....(((....)))...((((.((((.--((((....)))))))))))))))).))))))..)))- ( -41.10, z-score = -2.41, R) >droGri2.scaffold_15126 2830811 107 - 8399593 CCCUCGGUCUU-GGUUAGGCU-GGUUCACCUGUUUAUGCCAGAUUCCAGGCAUGCGAUUAAAAUCACCUGCGCAGGCA--UGCGCCCACGCAACCUUGCCUCU-----GUUGCCUG- .....(((((.-((((((((.-((....)).))))).)))))))).((((((((((..............))))((((--((((....))))....))))...-----..))))))- ( -32.24, z-score = -0.39, R) >consensus GCUUGAGUCUU_GGGAGGU__AGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGAAGUCUUGGAGUCGCAACUC_ .............................((((.((((((((....))))))))...............((((((....))))))....))))........................ (-19.66 = -19.99 + 0.33)

| Location | 15,368,875 – 15,368,989 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.68 |
| Shannon entropy | 0.52123 |
| G+C content | 0.54101 |
| Mean single sequence MFE | -44.35 |
| Consensus MFE | -31.00 |
| Energy contribution | -30.86 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15368875 114 - 23011544 GGAGUUGCGACUCCAAGACUUCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGUAAUCUGGCAUAAACAGGUGAACCU--ACCUCUC-AAGUCUCGAGC (((((....))))).((((((.(((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....)))--.....))-))))))..... ( -47.60, z-score = -4.38, R) >droSim1.chr2L 15127294 114 - 22036055 GGAGUUGCGACUCCAAGACUUCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCU--ACCUCUC-AAGUCUCGAGC (((((....))))).((((((.(((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....)))--.....))-))))))..... ( -47.30, z-score = -4.01, R) >droSec1.super_3 1629178 114 - 7220098 GGAGUUGCGACUCCAAGACUUCGACUGUGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCU--ACCUCUC-AAGUCUCGAGC (((((....))))).((((((.(((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....)))--.....))-))))))..... ( -45.40, z-score = -3.60, R) >droYak2.chr2L 2366674 114 + 22324452 GGAGUUGCGACUCCAAGACUUCUACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCU--ACCUCUC-AAGUCUCAAGC (((((....))))).((((((...(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....)))--.......-))))))..... ( -46.40, z-score = -4.29, R) >droEre2.scaffold_4929 7118505 114 - 26641161 GGAGUUGCGACUCCAAGACUUCCACUGCGUGGGCGCAAAAGCUUGCGCAGAUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCU--ACCUCUC-AAGUCUCAAGU (((((....))))).((((((.(((((((..(((......)))..))))).))...........(((((((....)))))))....(((....)))--.......-))))))..... ( -43.10, z-score = -3.64, R) >droAna3.scaffold_12916 7675661 95 - 16180835 ---------------GGACUGCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAA------CCACCG-AAGACUCCACC ---------------(((((.((.(((((..(((......)))..)))))(((((....)))))(((((((....))))))).....((....------))..))-.))..)))... ( -38.50, z-score = -3.65, R) >dp4.chr4_group4 4157381 102 + 6586962 --------GAGGCUGAGGCUGCGGCUGCGUGGGCGCAAAAGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAA------CCUCCG-AAGACUCCGAC --------....(.(((.((.((((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....------))))))-.)).))).).. ( -43.30, z-score = -2.82, R) >droPer1.super_10 1993426 108 + 3432795 --GAGGCUGCGGCUGCGGCUUCGGCUGCGUGGGCGCAAAAGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAA------CCUCCG-AAGACUCCGAC --((((((((....)))))((((((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....------))))))-))..))).... ( -50.50, z-score = -3.54, R) >droWil1.scaffold_180764 2214380 96 + 3949147 --------------------CCAACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCUCGCCUUACC-UCAACUCAACU --------------------....(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((((.....)))))....-........... ( -37.70, z-score = -4.22, R) >droMoj3.scaffold_6500 11603764 106 + 32352404 -GGGGCAAC--------UGUGAGGCUGCGUGGGCGCA--UGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCAAGUCUAACCGAAGACCCAGGG -((..((.(--------(((....(((((..(((...--.)))..)))))(((((....)))))(((((((....)))))))..)))).))..))..((((......))))...... ( -44.70, z-score = -2.86, R) >droVir3.scaffold_12963 8171817 113 + 20206255 -CAGGCAACUCCGAGGCUGCGAGGCUGCGUGGGCGCA--UGCCUGCGCAGGUGAUUUUAAUCGCAUGCCUGGAAUCUGGCAUAAACAGGUGAACCAAGUCUAACC-AAGACUGAGGG -((((((.........(((((((((((((....))))--.)))).)))))(((((....))))).))))))..(((((.......)))))...((.(((((....-.)))))..)). ( -46.90, z-score = -2.66, R) >droGri2.scaffold_15126 2830811 107 + 8399593 -CAGGCAAC-----AGAGGCAAGGUUGCGUGGGCGCA--UGCCUGCGCAGGUGAUUUUAAUCGCAUGCCUGGAAUCUGGCAUAAACAGGUGAACC-AGCCUAACC-AAGACCGAGGG -.((((...-----..(((((...(((((..(((...--.)))..)))))(((((....))))).)))))((.(((((.......)))))...))-.))))..((-........)). ( -40.80, z-score = -1.94, R) >consensus _GAGUUGCGACUCCAAGACUUCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCU__ACCUCCC_AAGACUCAAGC ........................(((((..(((......)))..)))))(((((....)))))(((((((....)))))))................................... (-31.00 = -30.86 + -0.14)
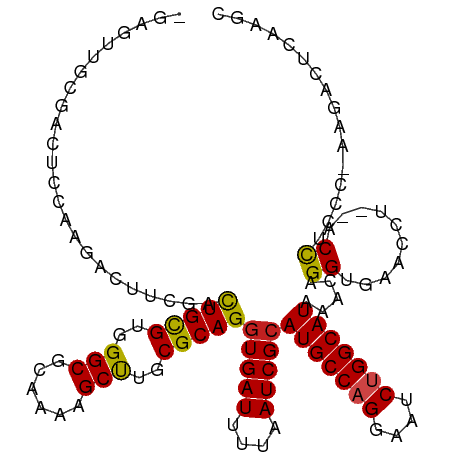
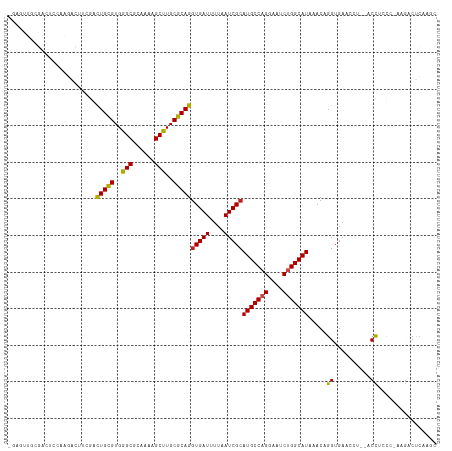
| Location | 15,368,876 – 15,368,990 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.97 |
| Shannon entropy | 0.51399 |
| G+C content | 0.53646 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -18.78 |
| Energy contribution | -18.96 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15368876 114 + 23011544 CUCGAGACUU-GA-GAGGUA-GGUUCACCUGUUUAUGCCAGAUUACUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGAAGUCUUGGAGUCGCAACUCCA ...(((((((-((-.(((((-((....)))...((((((((....))))))))...........))))((....))..(((((....))))))).)))))))(((((....))))). ( -39.50, z-score = -2.43, R) >droSim1.chr2L 15127295 114 + 22036055 CUCGAGACUU-GA-GAGGUA-GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGAAGUCUUGGAGUCGCAACUCCA ...(((((((-((-.(((((-((....)))...((((((((....))))))))...........))))((....))..(((((....))))))).)))))))(((((....))))). ( -39.90, z-score = -2.45, R) >droSec1.super_3 1629179 114 + 7220098 CUCGAGACUU-GA-GAGGUA-GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACACAGUCGAAGUCUUGGAGUCGCAACUCCA ...(((((((-((-.(((((-((....)))...((((((((....))))))))...........))))((((((....))))))........)).)))))))(((((....))))). ( -39.00, z-score = -2.52, R) >droYak2.chr2L 2366675 114 - 22324452 CUUGAGACUU-GA-GAGGUA-GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUAGAAGUCUUGGAGUCGCAACUCCA ...(((((((-..-.(((((-((....)))...((((((((....))))))))...........))))((....))..(((((....)))))...)))))))(((((....))))). ( -39.20, z-score = -2.42, R) >droEre2.scaffold_4929 7118506 114 + 26641161 CUUGAGACUU-GA-GAGGUA-GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCAUCUGCGCAAGCUUUUGCGCCCACGCAGUGGAAGUCUUGGAGUCGCAACUCCA ...(((((((-.(-((((((-((....)))...((((((((....))))))))........))).)))((((((....))))))((((...)))))))))))(((((....))))). ( -39.30, z-score = -2.00, R) >droAna3.scaffold_12916 7675662 94 + 16180835 GUGGAGUCUU-CG-GUGGU-----UCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGCAGUCC---------------- ((((......-.(-(((((-----(.....(((((((((((....)))))))).)))...))))))).((((((....))))))))))((....)).....---------------- ( -33.90, z-score = -2.58, R) >dp4.chr4_group4 4157382 101 - 6586962 UCGGAGUCUU-CG-GAGGU-----UCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCUUUUGCGCCCACGCAGCCGCAGCCUCAGCCUC--------- ...(((.((.-((-((((.-----...)))...((((((((....)))))))).............(((((..(((......)))..)))))))).)).)))......--------- ( -34.70, z-score = -1.41, R) >droPer1.super_10 1993427 107 - 3432795 UCGGAGUCUU-CG-GAGGU-----UCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCUUUUGCGCCCACGCAGCCGAAGCCGCAGCCGCAGCCUC--- .(((.(.(((-((-((((.-----...)))...((((((((....)))))))).............(((((..(((......)))..))))))))))).)....))).......--- ( -38.00, z-score = -1.18, R) >droWil1.scaffold_180764 2214381 95 - 3949147 GUUGAGUUGA-GGUAAGGCGAGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUUGG--------------------- .........(-(((...((.(((....)))....(((((((....)))))))))(((....)))))))((....))..(((((....)))))....--------------------- ( -27.70, z-score = -0.16, R) >droMoj3.scaffold_6500 11603765 105 - 32352404 CCUGGGUCUUCGGUUAGACUUGGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAGGCAU--GCGCCCACGCAGCCUCACA--------GUUGCCCC-- .(..(((((......)))))..)..((.((((.((((((((....)))))))).............(((((..(((..--..)))..)))))....)))--------).))....-- ( -35.30, z-score = -1.53, R) >droVir3.scaffold_12963 8171818 112 - 20206255 CCUCAGUCUU-GGUUAGACUUGGUUCACCUGUUUAUGCCAGAUUCCAGGCAUGCGAUUAAAAUCACCUGCGCAGGCAU--GCGCCCACGCAGCCUCGCAGCCUCGGAGUUGCCUG-- ...(((...(-((((((((..((....)).))))).))))((((((((((....(((....)))...((((.((((.(--(((....)))))))))))))))).))))))..)))-- ( -41.10, z-score = -2.42, R) >droGri2.scaffold_15126 2830812 106 - 8399593 CCUCGGUCUU-GGUUAGGCU-GGUUCACCUGUUUAUGCCAGAUUCCAGGCAUGCGAUUAAAAUCACCUGCGCAGGCAU--GCGCCCACGCAACCUUGCCUCU-----GUUGCCUG-- ....(((((.-((((((((.-((....)).))))).)))))))).((((((((((..............))))(((((--(((....))))....))))...-----..))))))-- ( -32.24, z-score = -0.38, R) >consensus CUUGAGUCUU_GG_GAGGUA_GGUUCACCUGUUUAUGCCAGAUUCCUGGCAUGCGAUUAAAAUCACCUGCGCAAGCUUUUGCGCCCACGCAGUCGAAGUCUUGGAGUCGCAACUC__ ............................((((.((((((((....))))))))...............((((........))))....))))......................... (-18.78 = -18.96 + 0.17)

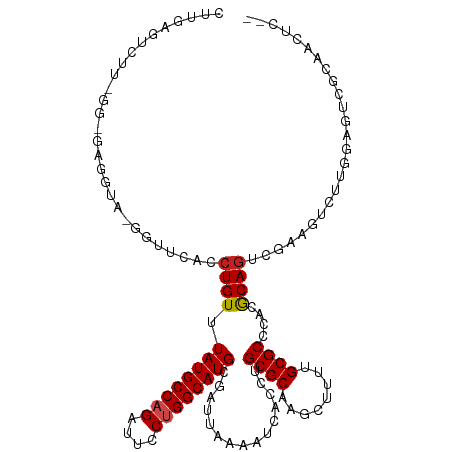
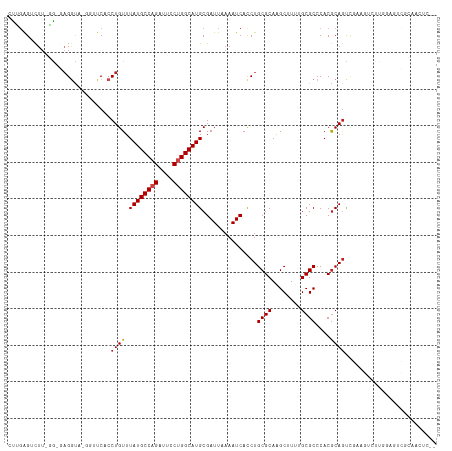
| Location | 15,368,876 – 15,368,990 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Shannon entropy | 0.51399 |
| G+C content | 0.53646 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -32.37 |
| Energy contribution | -31.95 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15368876 114 - 23011544 UGGAGUUGCGACUCCAAGACUUCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGUAAUCUGGCAUAAACAGGUGAACC-UACCUC-UC-AAGUCUCGAG ((((((....))))))((((((.(((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))-).....-))-)))))).... ( -48.30, z-score = -4.81, R) >droSim1.chr2L 15127295 114 - 22036055 UGGAGUUGCGACUCCAAGACUUCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC-UACCUC-UC-AAGUCUCGAG ((((((....))))))((((((.(((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))-).....-))-)))))).... ( -48.00, z-score = -4.42, R) >droSec1.super_3 1629179 114 - 7220098 UGGAGUUGCGACUCCAAGACUUCGACUGUGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC-UACCUC-UC-AAGUCUCGAG ((((((....))))))((((((.(((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))-).....-))-)))))).... ( -46.10, z-score = -3.97, R) >droYak2.chr2L 2366675 114 + 22324452 UGGAGUUGCGACUCCAAGACUUCUACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC-UACCUC-UC-AAGUCUCAAG ((((((....))))))((((((...(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((....))-).....-..-)))))).... ( -47.10, z-score = -4.70, R) >droEre2.scaffold_4929 7118506 114 - 26641161 UGGAGUUGCGACUCCAAGACUUCCACUGCGUGGGCGCAAAAGCUUGCGCAGAUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC-UACCUC-UC-AAGUCUCAAG ((((((....))))))((((((.(((((((..(((......)))..))))).))...........(((((((....)))))))....(((....))-).....-..-)))))).... ( -43.80, z-score = -3.98, R) >droAna3.scaffold_12916 7675662 94 - 16180835 ----------------GGACUGCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGA-----ACCAC-CG-AAGACUCCAC ----------------(((((.((.(((((..(((......)))..)))))(((((....)))))(((((((....))))))).....((...-----.))..-))-.))..))).. ( -38.50, z-score = -3.86, R) >dp4.chr4_group4 4157382 101 + 6586962 ---------GAGGCUGAGGCUGCGGCUGCGUGGGCGCAAAAGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGA-----ACCUC-CG-AAGACUCCGA ---------....(.(((.((.((((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((...-----.))))-))-.)).))).). ( -43.30, z-score = -2.62, R) >droPer1.super_10 1993427 107 + 3432795 ---GAGGCUGCGGCUGCGGCUUCGGCUGCGUGGGCGCAAAAGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGA-----ACCUC-CG-AAGACUCCGA ---((((((((....)))))((((((((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((...-----.))))-))-))..)))... ( -50.50, z-score = -3.32, R) >droWil1.scaffold_180764 2214381 95 + 3949147 ---------------------CCAACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCUCGCCUUACC-UCAACUCAAC ---------------------....(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....(((((.....)))))....-.......... ( -37.70, z-score = -4.35, R) >droMoj3.scaffold_6500 11603765 105 + 32352404 --GGGGCAAC--------UGUGAGGCUGCGUGGGCGC--AUGCCUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACCAAGUCUAACCGAAGACCCAGG --((..((.(--------(((....(((((..(((..--..)))..)))))(((((....)))))(((((((....)))))))..)))).))..))..((((......))))..... ( -44.70, z-score = -3.08, R) >droVir3.scaffold_12963 8171818 112 + 20206255 --CAGGCAACUCCGAGGCUGCGAGGCUGCGUGGGCGC--AUGCCUGCGCAGGUGAUUUUAAUCGCAUGCCUGGAAUCUGGCAUAAACAGGUGAACCAAGUCUAACC-AAGACUGAGG --((((((.........(((((((((((((....)))--).)))).)))))(((((....))))).))))))..(((((.......)))))...((.(((((....-.)))))..)) ( -45.60, z-score = -2.50, R) >droGri2.scaffold_15126 2830812 106 + 8399593 --CAGGCAAC-----AGAGGCAAGGUUGCGUGGGCGC--AUGCCUGCGCAGGUGAUUUUAAUCGCAUGCCUGGAAUCUGGCAUAAACAGGUGAACC-AGCCUAACC-AAGACCGAGG --.((((...-----..(((((...(((((..(((..--..)))..)))))(((((....))))).)))))((.(((((.......)))))...))-.))))....-.......... ( -39.80, z-score = -1.85, R) >consensus __GAGUUGCGACUCCAAGACUUCGACUGCGUGGGCGCAAAAGCUUGCGCAGGUGAUUUUAAUCGCAUGCCAGGAAUCUGGCAUAAACAGGUGAACC_UACCUC_CC_AAGACUCAAG .........................(((((..(((......)))..)))))(((((....)))))(((((((....)))))))....((((.......))))............... (-32.37 = -31.95 + -0.42)

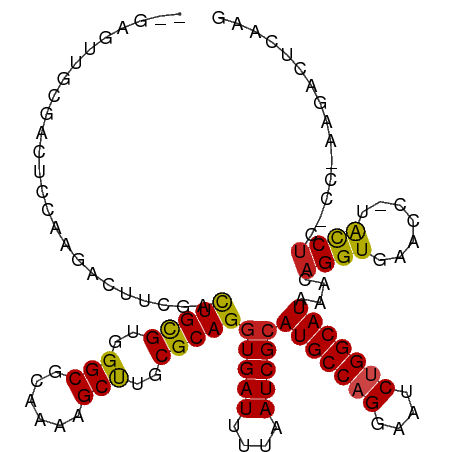
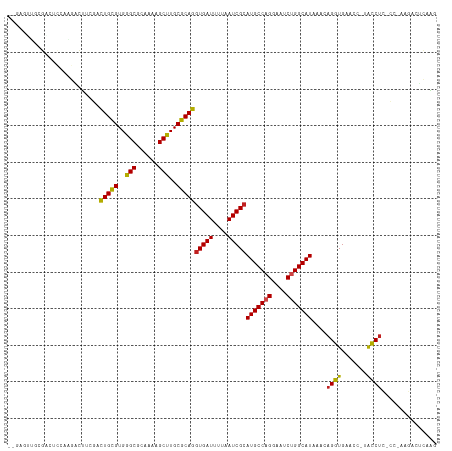
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:32 2011