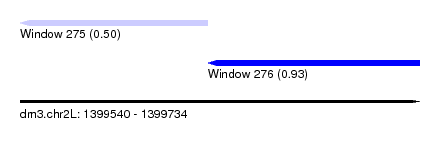
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,399,540 – 1,399,734 |
| Length | 194 |
| Max. P | 0.929473 |

| Location | 1,399,540 – 1,399,631 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 67.20 |
| Shannon entropy | 0.64340 |
| G+C content | 0.50132 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -12.20 |
| Energy contribution | -12.17 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 1399540 91 - 23011544 UGUCUCUUUCUGGCCCCCAAAUGUGAGCGCGAUUGUUUGCUUAGGUGCA----UGCUUGCAUGAUUUCAUCGCCGGCAGCACUUUUUGCCUCUGU---- ...........(((........(..((((....))))..)..((((((.----((((.(((((....))).)).))))))))))...))).....---- ( -24.70, z-score = -0.21, R) >droSim1.chr2L 1372816 91 - 22036055 UGUCUCUUUCUGGCCCCCAAAUGUGAGCGCGAUUGUUUGCUUAGGUGCA----UGCUUGCAUGAUUUCAUCGCCGGCAGCACUUUUUGCCUCUGU---- ...........(((........(..((((....))))..)..((((((.----((((.(((((....))).)).))))))))))...))).....---- ( -24.70, z-score = -0.21, R) >droSec1.super_14 1347233 91 - 2068291 UGUCUCUUUCUGGCCCCCAAAUGUGAGCGCGAUUGUUUGCUUAGGUGCA----UGCUUGCAUGAUUUCAUCGCCGGCAGCACUUUUUGCCUCUGU---- ...........(((........(..((((....))))..)..((((((.----((((.(((((....))).)).))))))))))...))).....---- ( -24.70, z-score = -0.21, R) >droEre2.scaffold_4929 1442688 91 - 26641161 CGUCUCUCUCUGGCCCCCAAAAGUGAGCGCGAUUGUUUGCUUAGGUGCA----UGCUUGCAUGAUUUCAUCGCCGGCAGCACUUUUUGCCUCUGU---- ...........(((......(((..((((....))))..)))((((((.----((((.(((((....))).)).))))))))))...))).....---- ( -27.00, z-score = -0.81, R) >droAna3.scaffold_12916 4861808 92 - 16180835 --CCCACAUAUGUGGGGAGUACGUAUACGCGU-UGUUUGCUUAGGUGCAU----GCUUGCAUGAUUUCAUCGCCGGCAGCACUCUUCGGGUGCCUCCGC --(((((....)))))..((((.((...(((.-....))).)).)))).(----(((.(((((....))).)).))))((((.(....)))))...... ( -31.80, z-score = -0.85, R) >dp4.chr4_group3 9916266 71 - 11692001 ---------------------UUCUGGCAUGUAUAUUUGCUUAGGUGCA---CUGCUUGCAUGCUUUCAACGCCAGCAGCACUUUUUGCUGCUGC---- ---------------------....((((((((....(((......)))---.....))))))))........((((((((.....)))))))).---- ( -25.20, z-score = -1.81, R) >droPer1.super_8 1123724 71 - 3966273 ---------------------UUCUGGCAUGUAUAUUUGCUUAGGUGCA---CUGCUUGCAUGCUUUCAACGCCAGCAGCACUUUUUGCUGCUGC---- ---------------------....((((((((....(((......)))---.....))))))))........((((((((.....)))))))).---- ( -25.20, z-score = -1.81, R) >droMoj3.scaffold_6500 28843120 77 + 32352404 ---------AGUUUCUGUAUGCAUAUGUGUGUCUGUUUGCAUAGGUGUAUGUGCGCUUGCAUGAUUUCAACGCCGGCAGCACUUUG------------- ---------.........((((((.((((((......)))))).))))))(((((((.((.((....))..)).))).))))....------------- ( -20.10, z-score = 0.46, R) >consensus __UCUCU_UCUGGCCCCCAAAUGUGAGCGCGAUUGUUUGCUUAGGUGCA____UGCUUGCAUGAUUUCAUCGCCGGCAGCACUUUUUGCCUCUGU____ .......................((((((........))))))(((((.....((((.((...........)).)))))))))................ (-12.20 = -12.17 + -0.03)

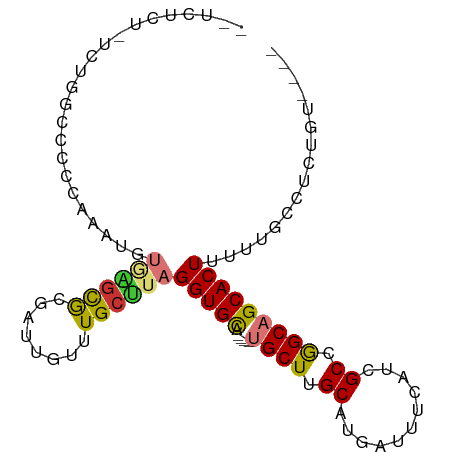
| Location | 1,399,631 – 1,399,734 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.50 |
| Shannon entropy | 0.23115 |
| G+C content | 0.59421 |
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.24 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1399631 103 - 23011544 GUUAGUGGCGCAUUAUGAGUUCAGCAGACACAUAUCGCUAUCCCUCUCUCUCGCU-CUCCCUCUGCGCCCCAUCUCGCCCCAUCUCUUUCCCCGCCACAACGUC (((.((((((......(((....(((((........((..............)).-.....)))))((........)).....)))......)))))))))... ( -20.18, z-score = -2.61, R) >droSim1.chr2L 1372907 99 - 22036055 GUUAGUGGCGCAUUAUGAGUUCAGCAGACACAAGCCCCUAUCCCCCUCUCUC--U-CACUCUCUGCGCCCCAUCUCG--CCAUCUCUCUUCCCGCCACAGCGCC (((.((((((......(((....(((((........................--.-.....)))))((........)--)...)))......)))))))))... ( -19.31, z-score = -1.84, R) >droEre2.scaffold_4929 1442779 101 - 26641161 GUUAGUGGCGCAUUAUGAGUUCAGCAGGCACAGACCCCUAUACCCGUCUCCCUCUGCGCCCUCUGCA-CCCAUCGCG--CCAUCUCUUUCUCCGCCACAGCGCC ....(((((((...(((.((.(((..(((.((((..................)))).)))..))).)-).))).)))--)))).........(((....))).. ( -28.27, z-score = -2.65, R) >consensus GUUAGUGGCGCAUUAUGAGUUCAGCAGACACAAACCCCUAUCCCCCUCUCUC_CU_CACCCUCUGCGCCCCAUCUCG__CCAUCUCUUUCCCCGCCACAGCGCC (((.((((((......(((....(((((.................................))))).................)))......)))))))))... (-16.68 = -16.24 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:29 2011