| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,306,317 – 15,306,411 |
| Length | 94 |
| Max. P | 0.789356 |

| Location | 15,306,317 – 15,306,411 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.59 |
| Shannon entropy | 0.69009 |
| G+C content | 0.47309 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -11.39 |
| Energy contribution | -10.68 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
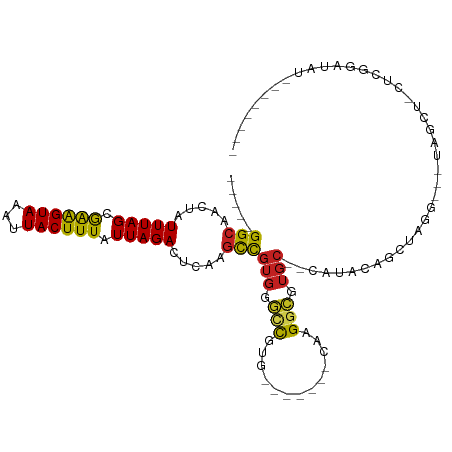
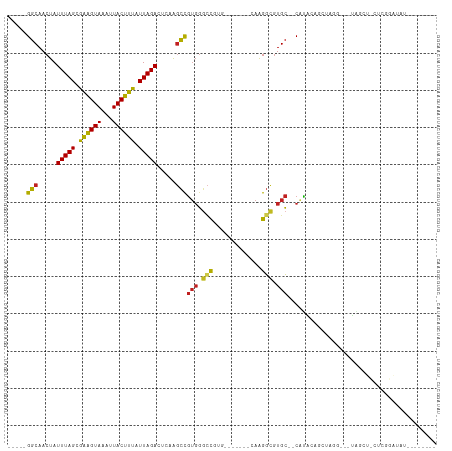
>dm3.chr2L 15306317 94 - 23011544 -----GGCAACUAUUUAGCGGAGUAAAUUACUUUAUUAGACUCGAGCCGUGGGCCGUG-------UAAGGCGUGC--CAUA----UAGC---UAGCUCCUCGGAUAUACAAAUCG -----(((.....(((((..(((((...)))))..))))).....)))(((..(((.(-------...(((..((--....----..))---..))).).)))...)))...... ( -20.20, z-score = 1.08, R) >droSim1.chr2L 15062744 89 - 22036055 -----GGCAACUAUUUAGCGGAGUAAAUUACUUUAUUAGACUCGAGCCGUGG--CGUG-------CAAGGCGUGC--CAUA----UAGC---UAGCU-CUCGGAUAUAUAUAU-- -----(((.....(((((..(((((...)))))..))))).....)))((((--((((-------(...))))))--))).----....---.....-...............-- ( -19.90, z-score = 0.86, R) >droSec1.super_3 1567596 92 - 7220098 -----GGCAACUAUUUAGCGGAGUAAAUUACUUUAUUAGACUCGAGCUGUGGGCCGUG-------CAAGGCGUGC--CAUA----UAGC---UAGCUCCUCGGAUAUAUAUAU-- -----(....)((((..(.(((((......((.....)).....(((((((((((((.-------....))).))--).))----))))---).))))))..)))).......-- ( -24.00, z-score = 0.04, R) >droYak2.chr2L 2301603 98 + 22324452 -----GGCAACUAUUUAGCGGAGUAAAUUACUUUAUUAGACUCAACCCGUGGGUCGUG-------GAAGGCGUGC--CAUAUAUGUAUG---UAGCUCCACGGAUAUUCAUACGA -----((((....(((.((((((((...))))))....((((((.....)))))))).-------)))....)))--).....((((((---....((....))....)))))). ( -24.10, z-score = -0.24, R) >droEre2.scaffold_4929 7054632 99 - 26641161 -----GGCAACUAUUUAGCGGAGUAAAUUACUUUAUUAGACUCCAGCCGUGGGCCGUGGAACGUUCAAGGCGUGCCACAUACUCGCAUG---UAGCUCCACGGAUAU-------- -----(....)........(((((.(((......)))..)))))..((((((((.((((.((((.....)))).)))).(((......)---)))).))))))....-------- ( -32.00, z-score = -2.00, R) >droAna3.scaffold_12916 7611601 81 - 16180835 -----GGCAACUAUUUAGCGGAGUAAAUUACUUUAUUAGACUCCGGCCGUGGGCUGUA-------CAAGGCGUGCCUCAUACAUGGAUA---UAGC------------------- -----((((.........((((((.(((......)))..))))))(((.((.......-------)).))).)))).............---....------------------- ( -20.00, z-score = -0.15, R) >dp4.chr4_group4 4093279 82 + 6586962 UUGCCGGCAACUAUUUAGUGAGGUAAAUUACUUUAUUAGAGUC-GGCCGUGGGCU--GU------CAAGGCGUGC--CCCGGAGCUCCA---CGGA------------------- ..((((((.....((((((((((((...)))))))))))))))-)))((((((((--.(------(..((....)--)..))))).)))---))..------------------- ( -31.10, z-score = -1.90, R) >droPer1.super_10 1929733 82 + 3432795 UUGCCGGCAACUAUUUAGUGAGGUAAAUUACUUUAUUAGAGUC-GGCCGUGGGCU--GU------CAAGGCGUGC--CCCGGAGCUCCA---CGGA------------------- ..((((((.....((((((((((((...)))))))))))))))-)))((((((((--.(------(..((....)--)..))))).)))---))..------------------- ( -31.10, z-score = -1.90, R) >droWil1.scaffold_180764 1835966 101 - 3949147 -----GGCAACUAUUUAGUGAGGUAAAUUACUUUAUUAGACUCUAGUCAGCCACCGUGGG--UUUUGGGUUUUGGGUCUUGGGGCUGGGAUCUGGGAUCUAGAGUCUG------- -----(((.((((((((((((((((...))))))))))))...))))..))).....((.--.(((((((((..((((((......))))))..)))))))))..)).------- ( -35.40, z-score = -2.75, R) >droVir3.scaffold_12963 11964266 90 - 20206255 -----GGCAACUAUUUAGUAAAGUAAAUUACUUUAUUAGAGGCAGGCUGUGGGCUU-GA------AAAGGCGUGC--CAUGCAGCUCAG---AUGCCUUGCAAGUGC-------- -----.((((...((((((((((((...))))))))))))((((((((((((((.(-(.------.....)).))--).)))))))...---.))))))))......-------- ( -30.60, z-score = -1.72, R) >droMoj3.scaffold_6500 20673729 91 - 32352404 -----GGCAACUAUUUAGUAAAGUAAAUUACUUUAUUAGAGGCAGGCCGUGGGCUUGUA------AAAGGCGUGC--CAUGCGGCUCAG---UUGUCUUGCAAGUGC-------- -----(((((((.((((((((((((...))))))))))))....((((((((((.(((.------....))).))--).))))))).))---)))))..........-------- ( -32.80, z-score = -2.72, R) >droGri2.scaffold_15126 5503364 90 - 8399593 -----GGCAACUAUUUAGUAAAGUAAAUUACUUUAUUAGAGGCAGGCCGUGGGCUU-CA------AAAGGCGUGC--CAUGCAGCUCAG---UUGGCUUGCAAGUGC-------- -----.(((.((.((((((((((((...)))))))))))).((((((((((((((.-((------...((....)--).)).)))))).---.)))))))).)))))-------- ( -33.30, z-score = -2.77, R) >consensus _____GGCAACUAUUUAGCGAAGUAAAUUACUUUAUUAGACUCAAGCCGUGGGCCGUG_______CAAGGCGUGC__CAUACAGCUAGG___UAGCU_CUCGGAUAU________ .....(((.....(((((.((((((...)))))).))))).....)))(((.(((.............))).)))........................................ (-11.39 = -10.68 + -0.71)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:26 2011