
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,298,550 – 15,298,665 |
| Length | 115 |
| Max. P | 0.697145 |

| Location | 15,298,550 – 15,298,653 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.23 |
| Shannon entropy | 0.81811 |
| G+C content | 0.52873 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -6.89 |
| Energy contribution | -6.27 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15298550 103 + 23011544 ---------CAAAACCAGCAAUCCAAC---CACCACC-CGGAUUCGAGAAUUGCAAAAUUUUUGCCCGCUUGAUAACCAACAGUGAUGGUAUGCCACUGGGUGGAGUCACAUUUGA---- ---------..................---..(((((-(((..(((((....((((.....))))...)))))..((((.......))))......))))))))............---- ( -25.80, z-score = -0.92, R) >droSim1.chr2L 15055117 103 + 22036055 ---------CAAAACCAAAAAUUCAUA---CUCCACC-CGGAUUCGAGAAUUGCAAAAUCUUUGCCCGCUUGAUAACCAACAGCGCUGGUAUGCCAGUGGGUGGAGUCACCUUUGA---- ---------.................(---(((((((-(((..(((((....((((.....))))...)))))...))......(((((....)))))))))))))).........---- ( -34.60, z-score = -3.87, R) >droSec1.super_3 1559905 103 + 7220098 ---------CAAAACCAAAAAUCAAUC---CUCCACC-CGGAUUCGAGAAUUGCAAAAUCUUUGCCCGCUUGAUAACCAACAGCGCUGGUAUGCCAGUGGGUGGAGUCACCUUUGA---- ---------..................---(((((((-(((..(((((....((((.....))))...)))))...))......(((((....)))))))))))))..........---- ( -33.50, z-score = -3.46, R) >droYak2.chr2L 2293699 103 - 22324452 ---------CAAAUACCACAAUCCAUC---CCCCACC-CGGCUUCGAGAAUUCCAAACUCUUUGCACGCCUAAUAACCAACAACACUGGUAUGCCCCUGGGUGGAGUCAACUUCGA---- ---------..................---..(((((-(((....(((.........)))...(((..((.................))..)))..))))))))............---- ( -21.73, z-score = -1.36, R) >droEre2.scaffold_4929 7046716 103 + 26641161 ---------UACCUAACAGAAUCCAUC---CGCCACC-UGGCUUCGAGAAUUCCAAACUCUUUGCACGCCUAACUACCAACAACACUGGUAUGCCACUGGGUGGCGUCAACUUCGA---- ---------.........(((......---(((((((-..(....(((.........)))...(((..((.................))..)))..)..))))))).....)))..---- ( -25.13, z-score = -2.12, R) >droAna3.scaffold_12916 7603677 108 + 16180835 ----AUUUCCAAAUCCCUUGAUUGGGC---CACCACC-UGGCUUCGAGUGCGCCAAGUUAUAUGCCCGCAUGAAGACCAAUCGCAUGGGCAUGCCCCUGGGUGGCGUGGACUUUGA---- ----.....((((((((((((...(((---((.....-)))))))))).(((((.......(((((((..(((.......)))..)))))))(((....))))))))))).)))).---- ( -37.40, z-score = -0.70, R) >dp4.chr4_group4 4086147 116 - 6586962 AGUCCCCCAAACGUCCCACGUUCAAACACGCACCGCCCUGGAUCCGACGGAGCCAGCAUCGGUGCCUCCGUCAUGACACAUUUCACGGGAAUGCCCGUGAGCUGCGUACUCUUUGG---- .........((((.....)))).......((((((..((((.(((...))).))))...))))))..(((.....((.((.((((((((....)))))))).)).))......)))---- ( -37.40, z-score = -1.31, R) >droPer1.super_10 1922567 116 - 3432795 AGUCCCCCAAACGUCCCACGUUCAAACACGCACCGCCCUGGAUCCGACGGAGCCAGCAUCGGUGCCUCCGUCAUGACACAUUUCACGGGAAUGCCCGUGAGCUGCGUACUCUUUGG---- .........((((.....)))).......((((((..((((.(((...))).))))...))))))..(((.....((.((.((((((((....)))))))).)).))......)))---- ( -37.40, z-score = -1.31, R) >droWil1.scaffold_180764 2103220 107 - 3949147 ---------CAUUUGCCACUUGUCCUG---CCCCACC-GGGAUUCGAAUCGGCAAAAAUGUAUGCACGUCUGCGUACCAAUGAUGAAGGUAUGCCUUUGGGCGGUGUUAAGCUGGAUAAG ---------......(((((((.((((---(((..((-((........)))).......(((((((....))))))).......(((((....))))))))))).).)))).)))..... ( -32.00, z-score = -0.62, R) >droVir3.scaffold_12963 11953474 104 + 20206255 --------ACAGCUACCAACAUUGGUC---CGCCACC-UGGCUUUGAGCACAUCAAAUUUUAUGGGCGACUGCGCACCAAUCGCAUGGGUGUGCCGCUGGGUGGCGUUACCUUUGA---- --------...........((..(((.---(((((((-..((...(.(((((((........((((((....))).)))........))))))))))..)))))))..)))..)).---- ( -43.19, z-score = -2.82, R) >droMoj3.scaffold_6500 20663923 103 + 32352404 ---------CGAUGCCGAACACUGGGC---CACCGCC-UGGGUACGAGAAUAUGAAAUUCUAUAGUCGCUUACGUACGAAUCGUUUGGGCGUGCCGCUGGGCGGCGUGACCUUCGA---- ---------(((..((((((.(..(((---....)))-..)(((((.((.((((......)))).)).....))))).....)))))).((((((((...))))))))....))).---- ( -33.30, z-score = 0.14, R) >droGri2.scaffold_15126 5494287 102 + 8399593 ----------GAAGUCCAACAUUGGUC---CGCCGCC-UGGCUAUGAGAAAUUCAAGUUCUACGGACGCUUGAAGACCAACCGCAUUGGUCUGCCGUUGGGCGGUGUGACCUUUGA---- ----------.........((..((((---(((((((-..((..........((((((((....)).))))))(((((((.....)))))))...))..))))))).))))..)).---- ( -46.60, z-score = -4.99, R) >consensus _________CAAAUCCCAAAAUCCAUC___CACCACC_UGGAUUCGAGAAUUCCAAAAUCUAUGCCCGCUUGAUAACCAACAGCACUGGUAUGCCACUGGGUGGCGUCACCUUUGA____ ................................(((((.((....)).....................................((.(((....))).)))))))................ ( -6.89 = -6.27 + -0.62)



| Location | 15,298,555 – 15,298,665 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.20 |
| Shannon entropy | 0.85322 |
| G+C content | 0.55601 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -6.89 |
| Energy contribution | -6.27 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.18 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15298555 110 + 23011544 ---CCAGCAAUCCAACCACCACC-CGGAUUCGAGAAUUGCAAAAUUUUUGCCCGCUUGAUAACCAACAGUGAUGGUAUGCCACUGGGUGGAGUCACAUUUGAGCUGGGCUCACC------ ---(((((..........(((((-(((..(((((....((((.....))))...)))))..((((.......))))......))))))))..(((....)))))))).......------ ( -34.90, z-score = -1.82, R) >droSim1.chr2L 15055122 110 + 22036055 ---CCAAAAAUUCAUACUCCACC-CGGAUUCGAGAAUUGCAAAAUCUUUGCCCGCUUGAUAACCAACAGCGCUGGUAUGCCAGUGGGUGGAGUCACCUUUGAGCUGGGCUCUCC------ ---............((((((((-(((..(((((....((((.....))))...)))))...))......(((((....)))))))))))))).......((((...))))...------ ( -38.00, z-score = -2.87, R) >droSec1.super_3 1559910 110 + 7220098 ---CCAAAAAUCAAUCCUCCACC-CGGAUUCGAGAAUUGCAAAAUCUUUGCCCGCUUGAUAACCAACAGCGCUGGUAUGCCAGUGGGUGGAGUCACCUUUGAGCUGGGCUCUCC------ ---(((....((((..(((((((-(((..(((((....((((.....))))...)))))...))......(((((....)))))))))))))......))))..))).......------ ( -37.10, z-score = -2.46, R) >droYak2.chr2L 2293704 110 - 22324452 ---ACCACAAUCCAUCCCCCACC-CGGCUUCGAGAAUUCCAAACUCUUUGCACGCCUAAUAACCAACAACACUGGUAUGCCCCUGGGUGGAGUCAACUUCGAGCUGGGCUCACC------ ---..................((-((((((.(((.((((((..(.....(((..((.................))..)))....)..))))))...))).))))))))......------ ( -29.83, z-score = -1.87, R) >droEre2.scaffold_4929 7046721 110 + 26641161 ---AACAGAAUCCAUCCGCCACC-UGGCUUCGAGAAUUCCAAACUCUUUGCACGCCUAACUACCAACAACACUGGUAUGCCACUGGGUGGCGUCAACUUCGAGCUGGGCUCACC------ ---....((..(((..(((((((-..(....(((.........)))...(((..((.................))..)))..)..)))))))((......))..)))..))...------ ( -29.73, z-score = -1.47, R) >droAna3.scaffold_12916 7603687 110 + 16180835 ---CCCUUGAUUGGGCCACCACC-UGGCUUCGAGUGCGCCAAGUUAUAUGCCCGCAUGAAGACCAAUCGCAUGGGCAUGCCCCUGGGUGGCGUGGACUUUGAACUCGUACCGCC------ ---.((......))((((((...-((((((((.(((.((..........)).))).))))).)))....((.(((....))).))))))))((((((.........)).)))).------ ( -36.80, z-score = -0.02, R) >dp4.chr4_group4 4086161 114 - 6586962 CCCACGUUCAAACACGCACCGCCCUGGAUCCGACGGAGCCAGCAUCGGUGCCUCCGUCAUGACACAUUUCACGGGAAUGCCCGUGAGCUGCGUACUCUUUGGAUGCGAGCCUAC------ .....((((......((((((..((((.(((...))).))))...)))))).((((.....((.((.((((((((....)))))))).)).))......))))...))))....------ ( -42.20, z-score = -2.38, R) >droPer1.super_10 1922581 114 - 3432795 CCCACGUUCAAACACGCACCGCCCUGGAUCCGACGGAGCCAGCAUCGGUGCCUCCGUCAUGACACAUUUCACGGGAAUGCCCGUGAGCUGCGUACUCUUUGGAUGCGAGCCUAC------ .....((((......((((((..((((.(((...))).))))...)))))).((((.....((.((.((((((((....)))))))).)).))......))))...))))....------ ( -42.20, z-score = -2.38, R) >droWil1.scaffold_180764 2103225 116 - 3949147 ---GCCACUUGUCCUGCCCCACC-GGGAUUCGAAUCGGCAAAAAUGUAUGCACGUCUGCGUACCAAUGAUGAAGGUAUGCCUUUGGGCGGUGUUAAGCUGGAUAAGAUGCCGCCGCCACC ---....(..((((((......)-)))))..)....(((......(((((((....))))))).......(((((....))))).(((((((((...........))))))))))))... ( -36.70, z-score = -0.28, R) >droVir3.scaffold_12963 11953480 110 + 20206255 ---ACCAACAUUGGUCCGCCACC-UGGCUUUGAGCACAUCAAAUUUUAUGGGCGACUGCGCACCAAUCGCAUGGGUGUGCCGCUGGGUGGCGUUACCUUUGACAUAACGCCGCC------ ---.....((..(((.(((((((-..((...(.(((((((........((((((....))).)))........))))))))))..)))))))..)))..)).............------ ( -43.19, z-score = -2.15, R) >droMoj3.scaffold_6500 20663928 101 + 32352404 ---CCGAACACUGGGCCACCGCC-UGGGUACGAGAAUAUGAAAUUCUAUAGUCGCUUACGUACGAAUCGUUUGGGCGUGCCGCUGGGCGGCGUGACCUUCGACAU--------------- ---.((.((.(..(((....)))-..))).))(((((.....)))))...(((((..(((..(((.....)))..)))(((((...)))))))))).........--------------- ( -31.80, z-score = 0.03, R) >droGri2.scaffold_15126 5494291 110 + 8399593 ---UCCAACAUUGGUCCGCCGCC-UGGCUAUGAGAAAUUCAAGUUCUACGGACGCUUGAAGACCAACCGCAUUGGUCUGCCGUUGGGCGGUGUGACCUUUGACGUCACACCGCC------ ---.(((((...((....))...-.(((..........((((((((....)).))))))(((((((.....)))))))))))))))((((((((((.......)))))))))).------ ( -48.40, z-score = -4.61, R) >consensus ___ACCAAAAUUCAUCCACCACC_UGGAUUCGAGAAUUCCAAAAUCUAUGCCCGCUUGAUAACCAACAGCACUGGUAUGCCACUGGGUGGCGUCACCUUUGAGCUGGGCCCGCC______ ..................(((((.((....)).....................................((.(((....))).))))))).............................. ( -6.89 = -6.27 + -0.62)

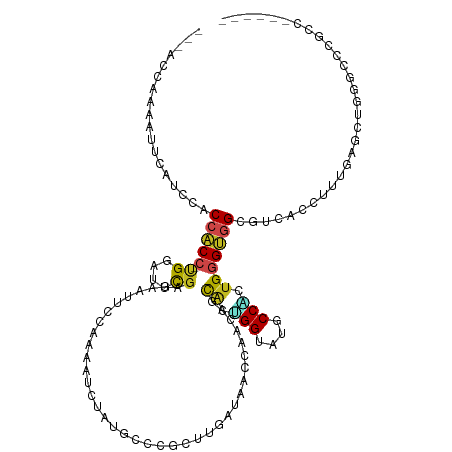
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:25 2011