
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,276,129 – 15,276,219 |
| Length | 90 |
| Max. P | 0.583725 |

| Location | 15,276,129 – 15,276,219 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.20 |
| Shannon entropy | 0.46591 |
| G+C content | 0.37114 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -10.07 |
| Energy contribution | -10.09 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583725 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 15276129 90 + 23011544 CUGCCC-GAAGCAUUUUGAUGUCGCAUUUUAGUGCAUUAAUUACUUUAU--GAUCCACUUUAUGGGCAUAAAAUAUGCAUGUUCCAAAAUCGG--------------- ....((-((.(((((..((((...))))..))))).........(((((--(.((((.....)))))))))).................))))--------------- ( -18.30, z-score = -0.74, R) >droSim1.chr2L 15032572 90 + 22036055 CUGCCC-GAAGCAUUUUGAUGUCGCAUUUUAGUGCAUUAAUUACUUUAU--GAUCCACUUUAUGGGCAUAAAAUAUGCAAGUUCCAAAAUCGG--------------- ....((-((.(((((..((((...))))..))))).........(((((--(.((((.....)))))))))).................))))--------------- ( -18.30, z-score = -0.89, R) >droSec1.super_3 1537826 90 + 7220098 UUGCCC-GAAGCAUUUUGAUGUCGCAUUUUAGUGCAUUAAUUACUUUAU--GAUCCACUUUAUGGGCAUAAAAUAUGCAAGUUCCAAAAUCGG--------------- ....((-((.(((((..((((...))))..))))).........(((((--(.((((.....)))))))))).................))))--------------- ( -18.30, z-score = -0.92, R) >droYak2.chr2L 2269656 90 - 22324452 CUGCCC-GAAGCAUUUUGAUGUCGCAUUAUAGUGCAUUAAUUACUUUAU--GAUCCACUUUAUGGGCAUAAAAUAUGCAAGUUCCAAAAUCGA--------------- .(((((-((((....((((((.(((......)))))))))...))))((--((......))))))))).........................--------------- ( -17.00, z-score = -0.56, R) >droEre2.scaffold_4929 7021032 90 + 26641161 CUGCCC-GAAGCAUUUUUAUGUCGCAUUAUAGUGCAUUAAUUACUUUAU--GAUCCACUUUAUGGGCAUAAAAUAUGCAAGUUUCAAAAUCGG--------------- ....((-((.(((((((((((((.(((...((((.((((.........)--))).))))..)))))))))))).))))..(...)....))))--------------- ( -20.40, z-score = -2.07, R) >droAna3.scaffold_12916 7577590 91 + 16180835 CUGCCCCGAAGCCUUUUCAUGUCGCAUUUUAGUGCAUUAAUUACUUUAU--GAUCCACUUUAUGCGCAUAAAAUAUGCAAGUCCCCCGGAUCG--------------- ......(((.((..(((.(((.(((((...((((.((((.........)--))).))))..)))))))).)))...))...(((...))))))--------------- ( -16.60, z-score = -1.41, R) >dp4.chr4_group4 4065881 92 - 6586962 CUGCCU-GGAGCAUUUUGAUGUCGCAUUUGAGUGCAUUAAUUACUUUAUAUGAUCCACUUUAUGAGCAUAAAAUAUGCACGCUCCGUCCCCGG--------------- ......-(((((...((((((.(((......)))))))))........(((((......))))).(((((...)))))..)))))........--------------- ( -20.50, z-score = -1.15, R) >droPer1.super_10 1902229 92 - 3432795 CUGCCU-GGAGCUUUUUGAUGUCGCAUUUGAGUGCAUUAAUUACUUUAUAUGAUCCACUUUAUGAGCAUAAAAUAUGCACGCUCCGUCCCCGG--------------- ......-(((((...((((((.(((......)))))))))........(((((......))))).(((((...)))))..)))))........--------------- ( -20.00, z-score = -1.19, R) >droMoj3.scaffold_6500 20631580 100 + 32352404 ----CCUGCGGCAUUUUAAUGUCGCAUUUUAGCGCAUUAAUUACUUUAU--GAUCCACUUUAUGCCCAUAAAAAAUGCGCAAAAAUAAAUAACAUAAAAUACCUGA-- ----...(((((((....)))))))(((((.(((((((..(((...(((--((......)))))....)))..))))))).)))))....................-- ( -21.30, z-score = -2.79, R) >droVir3.scaffold_12963 11927098 96 + 20206255 ----CCUGCAGCGUUUUAAUGUCGCAUUUUAGCGCAUUAAUUACUUUAU--GAUCCACUUUAUGCGCAUAAAAUAUGCGCAACGUUAAAU----UUAACCAAGCAA-- ----..(((...((.((((((.(((......)))))))))..)).....--...........((((((((...)))))))).........----........))).-- ( -22.20, z-score = -1.78, R) >droGri2.scaffold_15126 5471298 102 + 8399593 ----CCUGCAGCGUUUUAAUGUCGCAUUUUAGCGCAUUAAUUACUUUAU--GAUCCACUUUAUGCGCAUAAAAUAUGCGCACCAUAAACUGCCUUUAUUUUCAUAAAA ----...((((.((.((((((.(((......)))))))))..)).....--...........((((((((...)))))))).......))))................ ( -23.40, z-score = -2.73, R) >consensus CUGCCC_GAAGCAUUUUGAUGUCGCAUUUUAGUGCAUUAAUUACUUUAU__GAUCCACUUUAUGGGCAUAAAAUAUGCAAGUUCCAAAAUCGG_______________ .......((((....((((((.(((......)))))))))...))))..................((((.....)))).............................. (-10.07 = -10.09 + 0.02)
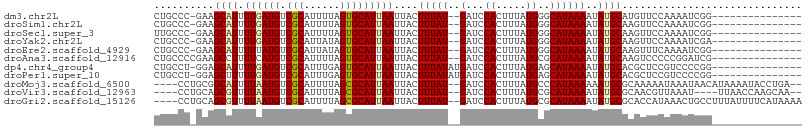
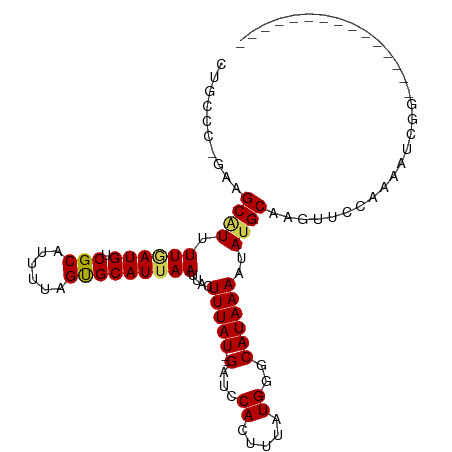
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:22 2011