| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,395,202 – 1,395,289 |
| Length | 87 |
| Max. P | 0.990679 |

| Location | 1,395,202 – 1,395,289 |
|---|---|
| Length | 87 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Shannon entropy | 0.48339 |
| G+C content | 0.39126 |
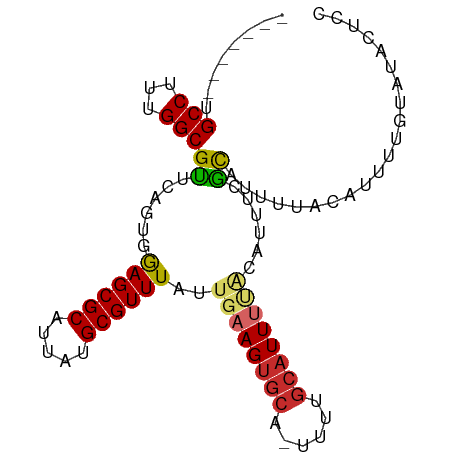
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990679 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
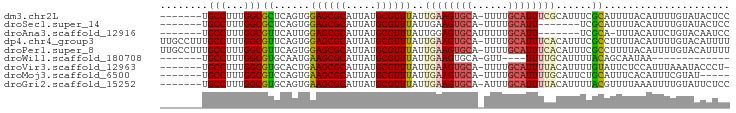
>dm3.chr2L 1395202 87 - 23011544 -------UGCCUUUGGCGCUCAGUGGAGCGCAUUAUGCGUUUAUUGAAGUGCA-UUUUGCAUUUCGCAUUUCGCAUUUUACAUUUUGUAUACUCC -------(((...((((((((....))))))...(((((.....((((((((.-....)))))))).....)))))....))....)))...... ( -26.10, z-score = -2.66, R) >droSec1.super_14 1343037 80 - 2068291 -------UGCCUUUGGCGCUCAGUGGAGCGCAUUAUGCGUUUAUUGAAGUGCA-UUUUGCAUU-------UCGCAUUUUACAUUUUGUAUACUCC -------.((.....((((((....)))))).....))((..((((((((((.-....)))))-------))).))...)).............. ( -22.10, z-score = -1.80, R) >droAna3.scaffold_12916 4857163 80 - 16180835 -------UGCCUUUGGCGUUCAUUGGAGCGCAUUAUGCGUUUAUUGGAGUGCAUUUUUGCAUU-------UCGCA-UUUACAUUCUGUACAAUCC -------.((.....((((((....)))))).....))((..(((((((((((....))))))-------))).)-)..)).............. ( -21.80, z-score = -1.82, R) >dp4.chr4_group3 9911411 94 - 11692001 UUGCCUUUGCCUUUGGCGUUCAGUGGAGCGCAUUAUGCGUUUAUUGAAGUGCA-UUUUGCAUUUCACAUUUCGCCUUUUACAUUUUGUACAUUUU .(((...((.....((((...((((((((((.....))))))..((((((((.-....)))))))))))).)))).....))....)))...... ( -25.30, z-score = -2.58, R) >droPer1.super_8 1118898 94 - 3966273 UUGCCUUUGCCUUUGGCGUUCAGUGGAGCGCAUUAUGCGUUUAUUGAAGUGCA-UUUUGCAUUUCACAUUUCGCCUUUUACAUUUUGUACAUUUU .(((...((.....((((...((((((((((.....))))))..((((((((.-....)))))))))))).)))).....))....)))...... ( -25.30, z-score = -2.58, R) >droWil1.scaffold_180708 3067762 70 - 12563649 -------UGCCUUUGGCGUGCAAUGAAGCGCAUUAUGCGUUUAUUGAAGUGCA-GUU----UUUUGCAUUUUACAGCAAUAA------------- -------.(((...))).(((....((((((.....))))))..(((((((((-(..----..))))))))))..)))....------------- ( -21.70, z-score = -1.88, R) >droVir3.scaffold_12963 7363428 86 - 20206255 -------UGCCUUUGGCGUGCACUGAAGCGCAUUAUGCGUUUAUUGAAGUGCA-UUUUGCAUUUUACAUUUUGUAUUCUCCAUUUAAAUACCCU- -------.(((...)))(((((((.((((((.....)))))).....))))))-)..((((..........))))...................- ( -19.20, z-score = -1.47, R) >droMoj3.scaffold_6500 28837502 82 + 32352404 -------UGCCUUUGGCGUCCAGUGAAGCGCAUUAUGCGUUUAUUGAAGUGCA-UUUUGCAUUUUGCAUUCUGCAUUUCACAUUUCGUAU----- -------........(((...((((((((((.....))))))..(((((((((-...(((.....)))...))))))))))))).)))..----- ( -24.70, z-score = -2.36, R) >droGri2.scaffold_15252 10492437 87 - 17193109 -------UGCCUUUGGCGUGCAGUGAAGCGCAUUAUGCGUUUAUUGAAGUGCA-AUUUGCAUUUUACAUUUUACGUUUUAAAUUUUGUAUUCUCC -------.......((((((.((((((((((.....))))))..((((((((.-....)))))))))))).)))))).................. ( -21.50, z-score = -1.75, R) >consensus _______UGCCUUUGGCGUUCAGUGGAGCGCAUUAUGCGUUUAUUGAAGUGCA_UUUUGCAUUUUACAUUUCGCAUUUUACAUUUUGUAUACUCC ........(((...)))((......((((((.....))))))..(((((((((....)))))))))......))..................... (-15.07 = -15.20 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:27 2011