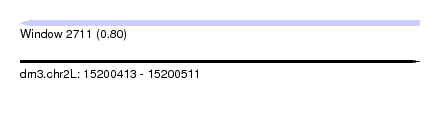
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,200,413 – 15,200,511 |
| Length | 98 |
| Max. P | 0.796997 |

| Location | 15,200,413 – 15,200,511 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.00 |
| Shannon entropy | 0.65719 |
| G+C content | 0.51537 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.31 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15200413 98 - 23011544 -----UGCCAU-AAAAUGCGCAUAAUGUGGCACGGAUGUGCCAGUCCACGUUGGUAAUUUGGCGCGUGAGCAUCG---CCGAUUAUCAU---UUUUGCCACAAGUGGCAG------- -----((((((-...((....))..(((((((..((((((((((......))))))..(((((((....))...)---))))....)))---)..))))))).)))))).------- ( -35.90, z-score = -1.80, R) >droSec1.super_3 1462709 101 - 7220098 -----UGCCAU-AAAAUGCGCAUAAUGUGGCACGGAUGUGCCAGUGCACGUUGGUAAUUUGGCGCGUGAGCAUCG---CCGAGUAUCGU---UCUUGCCACCAGUGGGGGCAG---- -----(((((.-...(((.((((....((((((....)))))))))).))))))))(((((((((....))...)---))))))...((---(((..(.....)..)))))..---- ( -38.30, z-score = -0.80, R) >droYak2.chr2L 2192926 97 + 22324452 -----UGCCAU-AAAAUGCGCAUAAUGUGGCACGGAUGUGUCAGUGCACGUUGGUAAUUUGGCGCGUGAGCAUCG---CCGAGUAUCGU---UCUUGCC-UCAGUGGCAG------- -----((((((-..((((.((((....((((((....)))))))))).))))(((((((((((((....))...)---)))))).....---...))))-...)))))).------- ( -34.80, z-score = -0.98, R) >droEre2.scaffold_4929 6943044 100 - 26641161 -----UGCCAU-AAAAUGCGCAUAAUGUGGCACGGAUGUGCCAGUGCACGUUGGUAAUUUGGCGCGUGAGCAUCG---CCGAGUAUCGUGUAUCUUGCC-CCAGUGGCAG------- -----((((((-..((((.((((....((((((....)))))))))).))))((((.((((((((....))...)---)))))))))............-...)))))).------- ( -37.30, z-score = -1.03, R) >droAna3.scaffold_12916 5500661 90 + 16180835 -----UGCCAU-AAAAUGCGCAUAAUGUGCCGCGGAUGUGCCAGUGCACGUUUGUUAUUUGGCGCGUGAGCAACGCUUU--GGGCUUAAGUGGAGCGG------------------- -----.((((.-.....((((.....)))).((((((((((....))))))))))....))))((....))..((((((--..((....)))))))).------------------- ( -33.10, z-score = -0.54, R) >dp4.chr4_group1 2895607 116 + 5278887 UGUCGUGCCAU-AAAAUCCGCAUAAUGUUGCACGGAUGUGUCAGUGCACGUUUGUUAUUUGGCGCGUGAGCACCGCUUUUGGGGUUGCACCGUGACGCCUCAGAUACGACGGGAUAG .(((((((((.-.......(((......)))((((((((((....))))))))))....))))((....))......((((((((..(.....)..)))))))).)))))....... ( -38.20, z-score = -0.13, R) >droWil1.scaffold_180764 3854267 80 - 3949147 -----UGCCAUAAAAAUCCGCAUAAUGUUGCACGGAUGUGUCAGUACACGUUUGCUAUUUGGCGGGUUAACAUCAAUCCCGGAAA-------------------------------- -----.((((.........(((......))).(((((((((....))))))))).....))))(((...........))).....-------------------------------- ( -20.70, z-score = -0.74, R) >droMoj3.scaffold_6500 24101194 90 + 32352404 -----GGCCAU-AAAAUGCGCAUAAUGUUGUGCGGAUGUGUCAGUACACGUUUGUUAUUUGGCGCGACAGCAGUUG-GCGGCAGCGGCAAGUCGCAG-------------------- -----.((((.-.....(((((......)))))((((((((....))))))))......))))(((((.((.((((-....)))).))..)))))..-------------------- ( -33.00, z-score = -1.16, R) >droGri2.scaffold_15252 1059979 84 - 17193109 -----GGCCAU-AAAAUGCGCAUAAUGUUUUGCGGAUGUGUCAGUACACGUUUGAUAUUUGGCGCGACAGUAGUUGCGGCGCAACGGUAG--------------------------- -----.(((..-....(((((...........(((((((((....)))))))))........((((((....)))))))))))..)))..--------------------------- ( -26.10, z-score = -0.64, R) >consensus _____UGCCAU_AAAAUGCGCAUAAUGUGGCACGGAUGUGUCAGUGCACGUUUGUUAUUUGGCGCGUGAGCAUCG___CCGAGUAUCGA___UCUUGCC_____U____________ ......((((.........((........))((.(((((((....))))))).))....))))((....)).............................................. (-16.41 = -16.31 + -0.10)

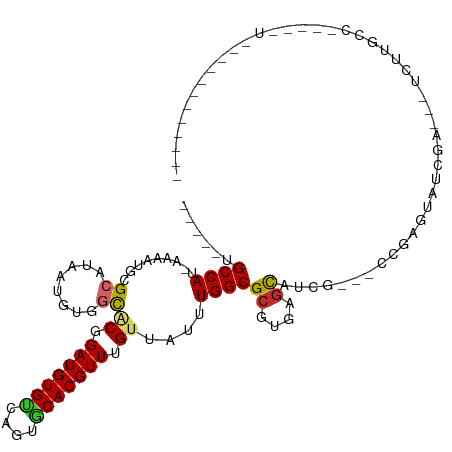
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:18 2011