| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,181,162 – 15,181,255 |
| Length | 93 |
| Max. P | 0.908433 |

| Location | 15,181,162 – 15,181,255 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.44 |
| Shannon entropy | 0.48547 |
| G+C content | 0.40237 |
| Mean single sequence MFE | -17.34 |
| Consensus MFE | -10.35 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15181162 93 - 23011544 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGGCAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGA---CUCUGC---GAGCCACUCCCACUUAAACU------ .((((((((((((...))))))((((((((............................)))))))).---....))---))))................------ ( -19.29, z-score = -1.93, R) >droSim1.chr2L 14930147 93 - 22036055 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGGCAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGA---CUCUGC---GAGCCACUCCCACUUAAACU------ .((((((((((((...))))))((((((((............................)))))))).---....))---))))................------ ( -19.29, z-score = -1.93, R) >droSec1.super_3 1443941 93 - 7220098 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGGCAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGA---CUCUGC---AAGCCACUCCCACUUAAACU------ .((.((.((((((...........)))))).))))............((((..........(((((.---..))))---)..........)))).....------ ( -15.55, z-score = -0.82, R) >droYak2.chr2L 2174065 93 + 22324452 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGGCAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGA---CUCUGC---GAGCCACUCCCACUUAAACU------ .((((((((((((...))))))((((((((............................)))))))).---....))---))))................------ ( -19.29, z-score = -1.93, R) >droEre2.scaffold_4929 6924449 93 - 26641161 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGGCAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGA---CUCUGC---GAGCCACUCCCACUUAAACU------ .((((((((((((...))))))((((((((............................)))))))).---....))---))))................------ ( -19.29, z-score = -1.93, R) >droAna3.scaffold_12916 5482764 81 + 16180835 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGACAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGA--CCCCAGU---G--CCACUG----------------- .((((((((((((...)))))..((((((...))))))..................))))).))...--...(((.---.--...)))----------------- ( -13.50, z-score = -0.97, R) >dp4.chr4_group1 2871626 102 + 5278887 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGACAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGAGACCUCUGU---GAUCCCCUCCUCCCACCGCACCAACC .(((((.((((((...........)))))).))).........................((((((((....)))))---.))).............))....... ( -16.60, z-score = -1.57, R) >droPer1.super_5 2795779 102 - 6813705 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGACAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGAGACCUCUGU---GAUCCCCUCCUCCCACCGCACCAUCC .(((((.((((((...........)))))).))).........................((((((((....)))))---.))).............))....... ( -16.60, z-score = -1.51, R) >droWil1.scaffold_180764 3830998 82 - 3949147 AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGGCAUAAAAAAUUAAAAGUAUAAAAACACUGGAACAACUUCA--CCCAAACC--------------------- .((.((.((((((...........)))))).))))..........................(((..........--.)))....--------------------- ( -11.20, z-score = -0.21, R) >droVir3.scaffold_12963 15819329 88 + 20206255 AGCUCGUUGACAUUUGAUGUCACUGUGUCACCGGCAUAAAAAAUUAAAAGUAUAAAAAUUAUGCAGA-UCCCAUGCCCAGAGUCAAGCC---------------- .(((((.((((((.((....))..)))))).)(((((............(((((.....))))).(.-...))))))..))))......---------------- ( -18.30, z-score = -1.99, R) >droMoj3.scaffold_6500 24073003 104 + 32352404 AGCUCGUUGACAUUUGAUGUCACUGUGUCAGCGGCAUAAAAAAUUAAAAGUAUUAAAAUGAUGCUGAAAUCUCAUGCCCAAGCCUUGGUCCACCCCCAAGUUUA- .(((.((((((((.((....))..))))))))(((((...........((((((.....))))))((....)))))))..)))(((((.......)))))....- ( -26.60, z-score = -2.95, R) >droGri2.scaffold_15252 1035840 84 - 17193109 AGCUCGUUGACAUUUGAUGUCACUGUGUCCCCGGCAUAAAAAAUUAAAAGUAUAAAAAUGAUGAAAA-CCCCAUGC---CAUGUAGCC----------------- .((((((.(((((.((....))..)))))...(((((............((......))........-....))))---)))).))).----------------- ( -12.55, z-score = -0.78, R) >consensus AGCUCGUUGACAUUUCAUGUCACUGUGUCACCGGCAUAAAAAAUUAAAAGUAUAAAAAUGAUGCAGA__CCUCUGC___GAGCCACUCCCACUUAAACU______ .((((((((((((...)))))..((((((...))))))..................))))).))......................................... (-10.35 = -10.50 + 0.15)

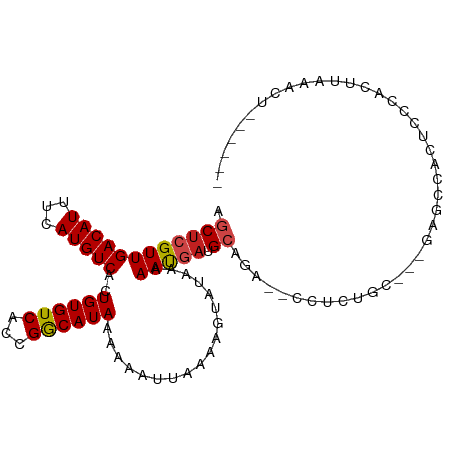
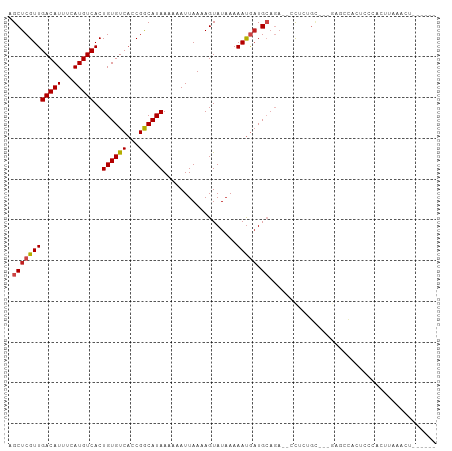
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:16 2011