| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,109,928 – 15,109,995 |
| Length | 67 |
| Max. P | 0.609016 |

| Location | 15,109,928 – 15,109,995 |
|---|---|
| Length | 67 |
| Sequences | 13 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 71.19 |
| Shannon entropy | 0.59737 |
| G+C content | 0.66674 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -12.43 |
| Energy contribution | -12.21 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.70 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
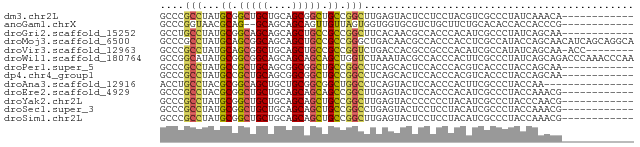
>dm3.chr2L 15109928 67 + 23011544 GCCCGCCUAUGCGGCUGCUGCAGCGGCUGCCGGCUUGAGUACUCCUCCUACGUCGCCCUAUCAAACA------------ ((..(((...((((((((....)))))))).)))..(((.....))).......))...........------------ ( -20.20, z-score = -0.22, R) >anoGam1.chrX 20278912 65 - 22145176 GCCCGGUAACGCAG--GCAGCAGCAGUUGUUAGUGGUGGUGCGUCUGCUUCUGCACACCACCACCCG------------ ...(((...(((..--(((((....)))))..)))((((((.((.(((....))).)))))))))))------------ ( -26.00, z-score = -1.20, R) >droGri2.scaffold_15252 952914 67 + 17193109 GCCUGCCUAUGCGGCAGCAGCAGCUGCCGCCGGCUUCACAACGCCACCCACAUCGCCCUAUCAGCAA------------ ...(((....((((((((....)))))))).(((........)))..................))).------------ ( -23.60, z-score = -2.13, R) >droMoj3.scaffold_6500 23962984 79 - 32352404 GCCCGCCUAUGCAGCGGCAGCAGCUGCCGCCGGGCUGACAACGCCACCCACCUCGCCAUACCAGCAACAUCAGCAGGCA (((.((...(((.(((((((...))))))).(((.((.......)))))..............)))......)).))). ( -26.30, z-score = -0.26, R) >droVir3.scaffold_12963 15721448 70 - 20206255 GCCCGCCUAUGCAGCGGCUGCAGCUGCCGCCGGUCUGACCACGCCGCCCACAUCGCCAUAUCAGCAA-ACC-------- ....((.(((((.(((((.......)))))((((........))))........).))))...))..-...-------- ( -16.50, z-score = 1.24, R) >droWil1.scaffold_180764 3740624 79 + 3949147 GCCGGCAUAUGCGGCGGCAGCAGCAGCAGCUGGUCUAAAUACGCCACCCACUUCGCCCUAUCAGCAGACCCAAACCCAA ...((((..((.((.(((..((((....))))((......))))).))))..).)))...................... ( -17.60, z-score = 1.29, R) >droPer1.super_5 2706803 67 + 6813705 GCCCGCCUAUGCCGCUGCAGCGGCGGCUGCCGGCCUCAGCACUCCACCCACGUCACCCUACCAGCAA------------ (((.((...((((((....))))))...)).))).................................------------ ( -18.90, z-score = 0.49, R) >dp4.chr4_group1 2785234 67 - 5278887 GCCCGCCUAUGCCGCUGCAGCGGCGGCUGCCGGCCUCAGCACUCCACCCACGUCACCCUACCAGCAA------------ (((.((...((((((....))))))...)).))).................................------------ ( -18.90, z-score = 0.49, R) >droAna3.scaffold_12916 5406533 64 - 16180835 ACCUGCCUACGCGGCAGCUGCUGCGGCGGCUGGCCUCAGUACUCCACCCACUUCGCCCUACCAA--------------- ....(((..((((((....))))))..))).(((...(((.........)))..))).......--------------- ( -19.10, z-score = 0.02, R) >droEre2.scaffold_4929 6850718 67 + 26641161 GCCCGCCUACGCGGCUGCUGCAGCAGCAGCCGGCUUGAGUACUCCACCCACAUCGCCCUACCAAACG------------ (((.((....))((((((((...))))))))))).................................------------ ( -19.90, z-score = -1.23, R) >droYak2.chr2L 2101661 67 - 22324452 GCCCGCCUAUGCGGCUGCUGCAGCAGCUGCCGGCUUGAGUACCCCCCCUACAUCGCCCUACCCAACG------------ ((..(((...((((((((....)))))))).)))..(((((.......))).))))...........------------ ( -20.40, z-score = -1.78, R) >droSec1.super_3 1371798 67 + 7220098 GCCCGCCUAUGCGGCUGCUGCAGCAGCUGCCGGCCUGAGUACUCCUCCUACAUCGCCCUACCAAACG------------ ((..(((...((((((((....)))))))).)))..(((.....))).......))...........------------ ( -20.50, z-score = -1.55, R) >droSim1.chr2L 14855146 67 + 22036055 GCCCGCCUAUGCGGCUGCUGCAGCAGCUGCCGGCUUGAGUACUCCUCCUACAUCGCCCUACCAAACG------------ ((..(((...((((((((....)))))))).)))..(((.....))).......))...........------------ ( -20.90, z-score = -1.64, R) >consensus GCCCGCCUAUGCGGCUGCUGCAGCAGCUGCCGGCCUGAGUACUCCACCCACAUCGCCCUACCAACAA____________ ....(((...((.(((((....))))).)).)))............................................. (-12.43 = -12.21 + -0.22)
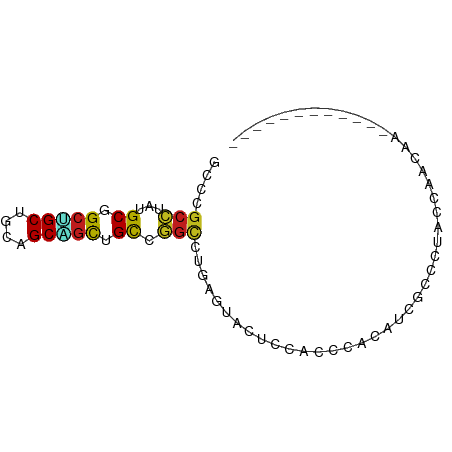

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:13 2011