
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,095,453 – 15,095,548 |
| Length | 95 |
| Max. P | 0.810846 |

| Location | 15,095,453 – 15,095,548 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 59.94 |
| Shannon entropy | 0.85492 |
| G+C content | 0.47601 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -11.81 |
| Energy contribution | -12.06 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.36 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
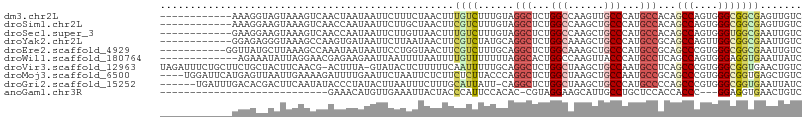
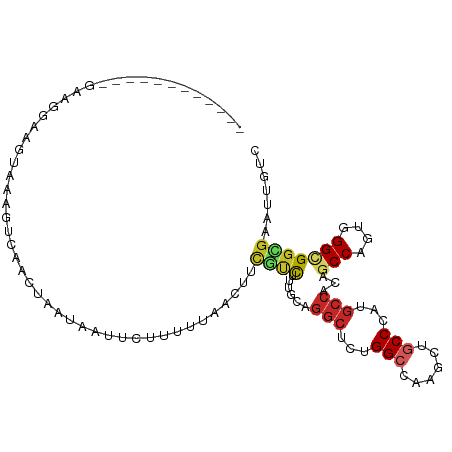
>dm3.chr2L 15095453 95 + 23011544 ------------AAAGGUAGUAAAGUCAACUAAUAAUUCUUUCUAACUUUGUCUUUGUAGGCUCUGGCCAAGUUGCCCAUGCCACAGCCAGUGGGCGGCGAGUUGUC ------------.....((((.......))))(((((((....................(((....)))..(((((((((((....))..)))))))))))))))). ( -25.30, z-score = -0.69, R) >droSim1.chr2L 14840991 95 + 22036055 ------------AAAGGAAGUAAAGUCAACCAAUAAUUCUUGCUAACUUCGUCUUUGUAGGCUCUGGCCAAGCUGCCCAUGCCACAGCCAGUGGGCGGCGAGUUGUC ------------...((..(......)..)).(((((((....................(((....)))..(((((((((((....))..)))))))))))))))). ( -27.90, z-score = -0.63, R) >droSec1.super_3 1357714 95 + 7220098 ------------GAAGGAAGUAAAGUCAACCAAUAAUUCUUGUUAACUUUGUCUUUGUAGGCUCUGGCCAAGCUGCCCAUGCCACAGCCAGUGGGUGGCGAAUUGUC ------------..(.((((((((((.(((...........))).)))))).)))).).(((....)))..((..(((((((....))..)))))..))........ ( -26.70, z-score = -0.72, R) >droYak2.chr2L 2080237 95 - 22324452 ------------GGAGAGGGUAAAGCCAAGUGAUAAUUCUUAAUAACUUCGUCUAUGCAGGCUCUGGCAAAGCUGCCCAUGCCGCAGCCAGUUGGCGGCGAAUUGUC ------------.(((((((.....)).(....)..))))).((((.((((((..(((.(((...((((....))))...))))))(((....))))))))))))). ( -27.80, z-score = 0.09, R) >droEre2.scaffold_4929 6836038 96 + 26641161 -----------GGUUAUGCUUAAAGCCAAAUAAUAAUUCCUGGUAACUUCGUCUUUGCAGGCUCUGGCAAAGCUGCCCAUGCCGCAGCCCGUGGGCGGCGAAUUGUC -----------((((........))))..............(..((.((((((..(((.(((...((((....))))...))))))(((....)))))))))))..) ( -29.60, z-score = -0.31, R) >droWil1.scaffold_180764 3709661 94 + 3949147 -------------AGAAAUAUUAGGAACGAGAAGAAUUAAUUUUAAUUUUGUUUUUUUAGGCACUGGCCAAGUUACCCAUGCCUCAGCCAGUGGGAGGUGAAUUAUC -------------.........((((((((((..((......))..)))))))))).....(((((((..((..........))..))))))).............. ( -16.60, z-score = 0.52, R) >droVir3.scaffold_12963 15698846 105 - 20206255 UAGAUUUCUGCUUCUGCUACUUCAACG-ACUUUA-GUAUACUCUUUUUCAAUUUUUGCAGGCUCUGGCUAAGCUGCCAAUGCCUCAGCCCGUGGGCGGUGAACUGUC ..((.....((....))....))....-....((-((.((((.............((.((((..((((......))))..))))))(((....))))))).)))).. ( -26.70, z-score = -0.66, R) >droMoj3.scaffold_6500 23937349 103 - 32352404 ----UGGAUUCAUGAGUUAAUUGAAAAGAUUUUGAAUUCUAAUUCUCUUCUCUUACCCAGGCUCUGGCUAAGCUGCCAAUGCCGCAGCCCGUGGGCGGUGAGCUGUC ----.(((((((.(((((.........))))))))))))............((((((..(((..((((......))))..)))...(((....)))))))))..... ( -31.80, z-score = -1.44, R) >droGri2.scaffold_15252 934748 100 + 17193109 ------UGAUUUGACACGACUUCAAUAUACCCUAUACUUAAUUUCUUUGCAUUAUU-CAGGCUCUGGCUAAGCUGCCCAUGCCCCAGCCCGUGGGCGGUGAAUUAUC ------((((((............................................-..(((....)))..((((((((((........)))))))))))))))).. ( -20.20, z-score = 0.27, R) >anoGam1.chr3R 5211962 75 - 53272125 ----------------------------GAAACAUGUUGAAAUUACUACCCAUUCCACAC-CGUAGGAAGCAUUGCCUGCUCCACCACCC---GGAGGUGAACUGUC ----------------------------.............................(((-((((((........)))))(((.......---)))))))....... ( -14.30, z-score = -0.04, R) >consensus ____________GAAGGAAGUAAAGUCAACUAAUAAUUCUUUUUAACUUCGUCUUUGCAGGCUCUGGCCAAGCUGCCCAUGCCACAGCCAGUGGGCGGCGAAUUGUC .................................................((((......(((...(((......)))...)))...(((....)))))))....... (-11.81 = -12.06 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:09 2011