| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,073,718 – 15,073,819 |
| Length | 101 |
| Max. P | 0.644415 |

| Location | 15,073,718 – 15,073,819 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 62.07 |
| Shannon entropy | 0.82977 |
| G+C content | 0.33031 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -5.26 |
| Energy contribution | -4.80 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.644415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
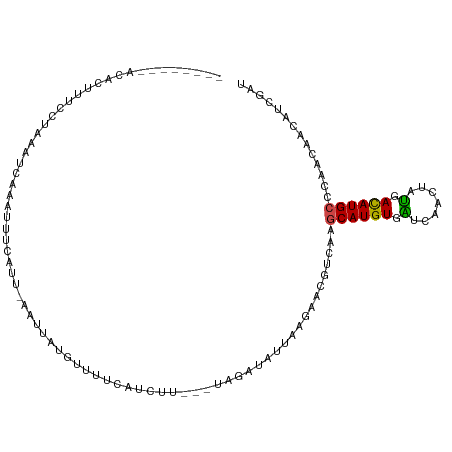
>dm3.chr2L 15073718 101 + 23011544 --------ACACUUUCCUGAAUCAAAUUUCAUUAAGUUCUUUUUUCAUCUUG-CUAGACAUUAAAAACAUCAAGCAUGUGAUCAACUAUGACAUGCCCAGCAAGAUCGAU --------..((((...((((......))))..)))).......((((((((-((.((...........))..((((((.((.....)).))))))..)))))))).)). ( -19.80, z-score = -2.39, R) >droSim1.chr2L 14818959 101 + 22036055 --------ACACGGUCCUGAAUCAAAUUUCAUUAAGUUCUUUUUUCAUCUUG-CUAGAUAUUAAAAACAUCAAGCAUGUGAUCAACUAUGACAUGCCCAAAAAUAUCGAU --------...((((..((((......))))........(((((..((((..-..))))...)))))......((((((.((.....)).))))))........)))).. ( -12.30, z-score = 0.13, R) >droSec1.super_3 1336159 100 + 7220098 --------ACACGUUCCUGAAUCAAAUUUCAUUAAGUUCUUUUU-CAUCUUG-CUAGAUAUUAAAAACAUCAAGCAUGUGAUCAACUAUGACAUGCCCAAAAAUAUCGAU --------.........((((......))))....(((.(((..-.((((..-..))))...)))))).....((((((.((.....)).)))))).............. ( -11.10, z-score = 0.14, R) >droYak2.chr2L 2050270 96 - 22324452 ---------UACUUUCCUAGAUUAAAUUUCUUAUAGUUGUGGUU-CAUCUC----AGAUAUCAAGAACAUCAAGCAUGUGGUCAACUUUGACAUGCCCAAUAACAUCGAU ---------..........................(((((.(((-(.....----.........)))).....((((((.(.......).))))))...)))))...... ( -12.64, z-score = 0.85, R) >droEre2.scaffold_4929 6813524 99 + 26641161 ---------UACUUUCCUAAAUCGAAUUUCCUACAAUUGUGUUU-CAUCUUG-CUAGAUAUCAAGAACAUCAAUCAUGUGGUCAACUUUGAUAUGCCCAACAACAUCGAU ---------...........((((((((..((((((((((((..-..(((((-........)))))))).))))..)))))..)).)))))).................. ( -13.10, z-score = -0.12, R) >droAna3.scaffold_12916 5367596 102 - 16180835 AUUCUUCGCUGAUUACCU--UUGGUAAUCCUCC--AUUAUUCUUGC-UCCU---UAGAUAUCAAGAAUGUCAAGCAUGUGGUUAACUUUGAUAUGCCCAACAACAUUGCC .......((.(((((((.--..)))))))....--..((((((((.-((..---..))...))))))))....((((((.(.......).))))))...........)). ( -17.80, z-score = -0.80, R) >dp4.chr4_group1 2733064 102 - 5278887 ---AAUGAAUAUUCAGAUAAUUUACGUUCCAUU--UUGAUGCAUUUCUGAU---CAGAUAUUAAGAACGUCAAGCAUGUCGUUAACUACGAUAUGCCCAAGACCAUUGAU ---.........((((.......((((((...(--(((((.((....))))---))))......))))))...(((((((((.....))))))))).........)))). ( -22.50, z-score = -2.57, R) >droPer1.super_5 2654802 102 + 6813705 ---AAUGAAUAUUCAGAUAAUUUACGUUCCAUU--UUGAUGCAUUUCUGAU---CAGAUAUUAAGAACGUCAAGCAUGUCGUUAACUACGAUAUGCCCAAGACCAUUGAU ---.........((((.......((((((...(--(((((.((....))))---))))......))))))...(((((((((.....))))))))).........)))). ( -22.50, z-score = -2.57, R) >droWil1.scaffold_180764 3653502 95 + 3949147 --------AUAUUUUUCC--UUGUUAUUUGUUA--AUUUUUGUUUUGUUUU---UAGACAUUAAGAACGUUAGUCAUGUCAUUAAUUAUGAUAUGCCCAGCACAAUAGAU --------..........--.....(((((((.--...........(((((---(((...))))))))(((.(.((((((((.....)))))))).).)))..))))))) ( -15.20, z-score = -1.71, R) >droMoj3.scaffold_6500 23904915 104 - 32352404 ------CUCCAGCUGUUUGUCUAUUUUUUGUUUAAACUAAAUCCCUCCUUUGGUUAGAUAUUAAAAAUGUGAAGCAUGUGAUUAACUAUGACAUGCCCUCAAAUAUCGAU ------.....(.((((((((((((((((((((((.(((((.......))))))))))))..))))))).)).((((((.((.....)).))))))...)))))).)... ( -19.10, z-score = -2.22, R) >droGri2.scaffold_15252 903223 109 + 17193109 UCUCUAAUCCAA-UAUAAAUGCACCAACUUAUUGUUGUAUCUCCACCACUCCACCAGAUAUAAAGAAUGUUAAGCAUGUGAUUAACUAUGACAUGCCAAAGACUGUGGAU ......(((((.-............(((......((((((((.............)))))))).....)))..((((((.((.....)).)))))).........))))) ( -18.32, z-score = -0.92, R) >droVir3.scaffold_12963 15660629 101 - 20206255 --------CCACUGCCCAAAAUUAUAUUCAAACUAAUCAUGUUGCAUCCUUU-GCAGAUAUUAAGAAUGUAAAGCAUGUUAUCAACUAUGAUAUGCCCACCAAUAUUGAU --------........(((..((((((((....((((....(((((.....)-))))..)))).)))))))).(((((((((.....))))))))).........))).. ( -17.50, z-score = -1.76, R) >consensus ________ACACUUUCCUAAAUCAAAUUUCAUU_AAUUAUGUUUUCAUCUU___UAGAUAUUAAGAACGUCAAGCAUGUGAUCAACUAUGACAUGCCCAACAACAUCGAU .........................................................................((((((.(.......).)))))).............. ( -5.26 = -4.80 + -0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:06 2011