| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,064,058 – 15,064,153 |
| Length | 95 |
| Max. P | 0.998416 |

| Location | 15,064,058 – 15,064,153 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Shannon entropy | 0.38925 |
| G+C content | 0.37104 |
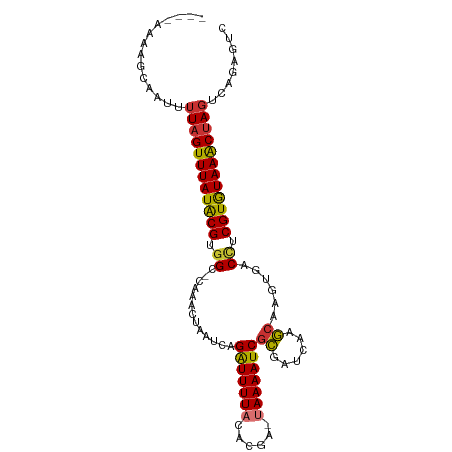
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.998416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
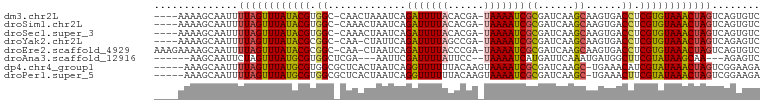
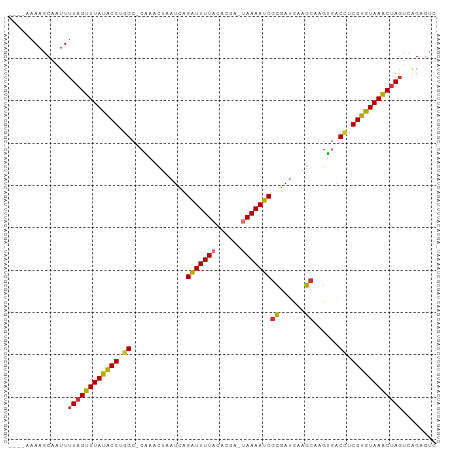
>dm3.chr2L 15064058 95 - 23011544 ----AAAAGCAAUUUUAGUUUAUACGUGGC-CAACUAAAUCAGAUUUUACACGA-UAAAAUCGCGAUCAAGCAAGUGACCUCGUGUAAACUAGUCAGUGUC ----....(((...((((((((((((.((.-..(((......(((((((.....-)))))))((......)).)))..)).))))))))))))....))). ( -26.50, z-score = -4.13, R) >droSim1.chr2L 14812650 95 - 22036055 ----AAAAGCAAUUUUAGUUUAUACGUGGC-CAAACUAAUCAGAUUUUACACGA-UAAAAUCGCGAUCAAGCAAGUGACCUCGUGUAAACUAGUCAGUGUC ----....(((...((((((((((((.((.-((.........(((((((.....-)))))))((......))...)).)).))))))))))))....))). ( -25.80, z-score = -3.87, R) >droSec1.super_3 1326242 95 - 7220098 ----AAAAGCAAUUUUAGUUUAUACGUGGC-CAAACUAAUCAGAUUUUACACGA-UAAAAUCGCGAUCAAGCAAGUGACCUCGUGUAAACUAGUCAGUGUC ----....(((...((((((((((((.((.-((.........(((((((.....-)))))))((......))...)).)).))))))))))))....))). ( -25.80, z-score = -3.87, R) >droYak2.chr2L 2040670 94 + 22324452 ----AAAAGCAAUUUUAGUUUAUACGCGGC-CAA-CUAUUCAGAUUUUAGCCGA-UAAAAUCGCGAUCAAGCAAGUGACCUCGUGUAAACUAGUCAGAGUC ----..........((((((((((((.((.-..(-((.....(((((((.....-)))))))((......)).)))..)).))))))))))))........ ( -25.30, z-score = -3.49, R) >droEre2.scaffold_4929 6803959 98 - 26641161 AAAGAAAAGCAAUUUUAGUUUAUACGCGGC-CAA-CUAAUCAGAUUUUACCCGA-UAAAAUCGCGAUCAAGCAAGUGACCUCGUGUAAACUAGUCAGUGUC ........(((...((((((((((((.((.-..(-((.....(((((((.....-)))))))((......)).)))..)).))))))))))))....))). ( -26.20, z-score = -3.99, R) >droAna3.scaffold_12916 5358635 87 + 16180835 ------AAGCAAUUCUAGUUUAUGCGUGGCUCGA---AAUUCGAUUUUAUUCC--UAAAAUCAUGAUUCAAAUGAUGGCUUCGUAUAAGCAA---AGAGUC ------.....(((((.(((((((((.((((((.---....))).........--....(((((.......))))).))).)))))))))..---))))). ( -20.70, z-score = -2.22, R) >dp4.chr4_group1 2723308 95 + 5278887 -----AAAGCAAUUUUAGUUUAUGCGUGGCGCUCACUAAUCAGGUUUUUUACAAGUAAAAUCGCGAUCAAGC-UGAAACAUCGUAUAAACUAGUCGGAAGA -----.........((((((((((((((....(((((.(((.((((((........))))))..)))..)).-)))..)).))))))))))))((....)) ( -21.30, z-score = -1.39, R) >droPer1.super_5 2645065 95 - 6813705 -----AAAGCAAUUUUAGUUUAUGCGUGGCGCUCACUAAUCAGGUUUUUUACAAGUAAAAUCGCGAUCAAGC-UGAAACUUCGUAUAAACUAGUCGGAAGA -----.........((((((((((((.((...(((((.(((.((((((........))))))..)))..)).-)))..)).))))))))))))((....)) ( -21.40, z-score = -1.22, R) >consensus ____AAAAGCAAUUUUAGUUUAUACGUGGC_CAAACUAAUCAGAUUUUACACGA_UAAAAUCGCGAUCAAGCAAGUGACCUCGUGUAAACUAGUCAGAGUC ..............((((((((((((.((.............(((((((......)))))))((......))......)).))))))))))))........ (-19.41 = -19.03 + -0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:04 2011