| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,039,296 – 15,039,402 |
| Length | 106 |
| Max. P | 0.627500 |

| Location | 15,039,296 – 15,039,402 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 56.55 |
| Shannon entropy | 0.87439 |
| G+C content | 0.28485 |
| Mean single sequence MFE | -14.40 |
| Consensus MFE | -5.68 |
| Energy contribution | -5.41 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.53 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589217 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 15039296 106 + 23011544 CUUUAACAGUGAUGUGAUGGGAAUAAAUAUACAAAUGUAUUCAGAAACUGCAAAGUUAAAACAAAAUAUAUUAGCUACUCCAAAUUGGCUAUAAAAUAACCAAAUG .....(((....)))..((((((((.((......)).)))))...((((....))))..............((((((........))))))........))).... ( -12.80, z-score = -0.00, R) >droPer1.super_5 2618256 101 + 6813705 CUUAUUCAGCGCCCUAUACUAAAUAAUU--AGAUUUGUAUU---AAAAUAUUCAGAAGUGACAGUAUAUCGCCACAAAAAACAUUUAGCUAUAAAAUAGCUGGCUC .......(((((..((((((..((((..--....))))...---.......(((....))).))))))..)).............(((((((...)))))))))). ( -13.10, z-score = 0.12, R) >dp4.chr4_group1 2691037 101 - 5278887 CUUAUUCAGCACCCUAUACUAAAUAGUU--AGAUUUGUAUU---AAAAUAUUCAGAAGUGACAGUAUAUCGCCACAAAAAACAUUUAGCUAUAAAAUAGCUGCCUC ......((((....((((((((((.(((--...((((.((.---.....)).)))).(((.(........).)))....))))))))).)))).....)))).... ( -11.90, z-score = -0.00, R) >droAna3.scaffold_12916 15358748 84 - 16180835 --------------------UUUUCGAUAAACAAGGGUUUAAAAAUGUAGUAAAAUAGUAACAUUUUGAUGAGAAUAUAAAUGAUGGGACCUAAAAUACCCUGG-- --------------------((((((.(((((....)))))(((((((............)))))))..))))))..........(((..........)))...-- ( -9.20, z-score = 0.87, R) >droEre2.scaffold_4929 6779024 106 + 26641161 CUUUUACAGUAAUGUGAGGAGAUCAUAUAAAAAAAUGUAUUCAGAAUUUGCUAAAUAGAAACGAAAAAUAUUAGCUUAUCCAAUUUGGCUAUAAUAUAGCCAAGUG ...(((((....)))))(((((..((((......))))..)).......(((((................)))))...))).((((((((((...)))))))))). ( -21.59, z-score = -2.57, R) >droSec1.super_3 1301437 106 + 7220098 CUUUUACAGUAAUAUGCAAAGAAUAGAUGUAAAAGUGUAUUCAGAAACUACUAAAUAAAAACAAAAAAUAUUAGCUACGCCAAGUUGGCUAUAAAAUAGCCAAAUG ((((((((....(((.......)))..)))))))).................................................((((((((...))))))))... ( -15.20, z-score = -0.90, R) >droSim1.chr2L 14786368 106 + 22036055 CUUUCAUAUUAAUAUGAUAUGAAUAGAUGUACAAGUGUAUUCAGAAACUACUAAAUAAAAGCAAAAAAUAUUAGCUACGCCAAGUUGGCUAUAAAAUAGCCAAAUG ..((((((((.....))))))))..((.((((....)))))).................(((...........)))........((((((((...))))))))... ( -17.00, z-score = -1.25, R) >consensus CUUUUACAGUAAUAUGAAAUGAAUAGAUAUACAAGUGUAUUCAGAAACUACUAAAUAGAAACAAAAUAUAUUAGCUAAACCAAUUUGGCUAUAAAAUAGCCAAAUG ....................................................................................((((((((...))))))))... ( -5.68 = -5.41 + -0.26)

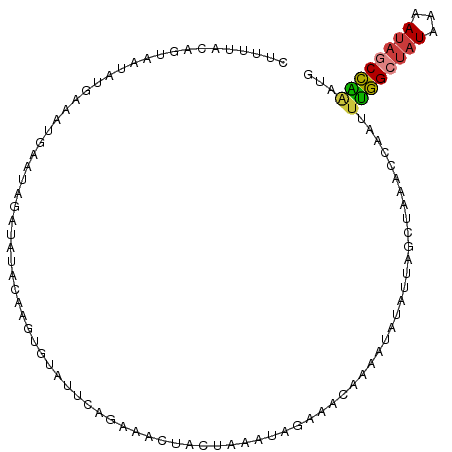

| Location | 15,039,296 – 15,039,402 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 56.55 |
| Shannon entropy | 0.87439 |
| G+C content | 0.28485 |
| Mean single sequence MFE | -17.04 |
| Consensus MFE | -5.20 |
| Energy contribution | -4.47 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627500 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 15039296 106 - 23011544 CAUUUGGUUAUUUUAUAGCCAAUUUGGAGUAGCUAAUAUAUUUUGUUUUAACUUUGCAGUUUCUGAAUACAUUUGUAUAUUUAUUCCCAUCACAUCACUGUUAAAG ...((((((((...))))))))...((((((...(((((((..(((...((((....)))).......)))...)))))))))))))................... ( -15.80, z-score = -0.68, R) >droPer1.super_5 2618256 101 - 6813705 GAGCCAGCUAUUUUAUAGCUAAAUGUUUUUUGUGGCGAUAUACUGUCACUUCUGAAUAUUUU---AAUACAAAUCU--AAUUAUUUAGUAUAGGGCGCUGAAUAAG ..(((((((((...))))))((((((((...((((((......))))))....)))))))).---.((((((((..--....)))).))))..))).......... ( -20.60, z-score = -1.06, R) >dp4.chr4_group1 2691037 101 + 5278887 GAGGCAGCUAUUUUAUAGCUAAAUGUUUUUUGUGGCGAUAUACUGUCACUUCUGAAUAUUUU---AAUACAAAUCU--AACUAUUUAGUAUAGGGUGCUGAAUAAG ....((((.((((((((.(((((((((.((((((..((.(((...(((....))).))).))---..))))))...--))).))))))))))))))))))...... ( -21.20, z-score = -1.23, R) >droAna3.scaffold_12916 15358748 84 + 16180835 --CCAGGGUAUUUUAGGUCCCAUCAUUUAUAUUCUCAUCAAAAUGUUACUAUUUUACUACAUUUUUAAACCCUUGUUUAUCGAAAA-------------------- --..(((((......(....)..................(((((((............)))))))...))))).............-------------------- ( -8.00, z-score = 0.23, R) >droEre2.scaffold_4929 6779024 106 - 26641161 CACUUGGCUAUAUUAUAGCCAAAUUGGAUAAGCUAAUAUUUUUCGUUUCUAUUUAGCAAAUUCUGAAUACAUUUUUUUAUAUGAUCUCCUCACAUUACUGUAAAAG ...((((((((...)))))))).(..((...(((((................)))))....))..)........(((((((((((........)))).))))))). ( -16.09, z-score = -1.37, R) >droSec1.super_3 1301437 106 - 7220098 CAUUUGGCUAUUUUAUAGCCAACUUGGCGUAGCUAAUAUUUUUUGUUUUUAUUUAGUAGUUUCUGAAUACACUUUUACAUCUAUUCUUUGCAUAUUACUGUAAAAG ...((((((((......(((.....))))))))))).............(((((((......)))))))..((((((((...................)))))))) ( -18.51, z-score = -1.61, R) >droSim1.chr2L 14786368 106 - 22036055 CAUUUGGCUAUUUUAUAGCCAACUUGGCGUAGCUAAUAUUUUUUGCUUUUAUUUAGUAGUUUCUGAAUACACUUGUACAUCUAUUCAUAUCAUAUUAAUAUGAAAG ...((((((((......(((.....))))))))))).........(((((((((((((.....((((((............)))))).....)))))).))))))) ( -19.10, z-score = -1.52, R) >consensus CAUUUGGCUAUUUUAUAGCCAAAUUGGUGUAGCUAAUAUAUUUUGUUUCUAUUUAGUAGUUUCUGAAUACACUUGUAUAUCUAUUCACUUCACAUUACUGUAAAAG .....((((((...))))))...................................................................................... ( -5.20 = -4.47 + -0.73)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:01 2011