
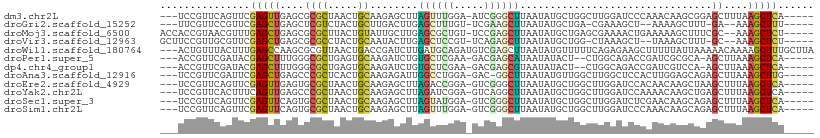
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,035,405 – 15,035,505 |
| Length | 100 |
| Max. P | 0.809198 |

| Location | 15,035,405 – 15,035,505 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 64.18 |
| Shannon entropy | 0.75356 |
| G+C content | 0.47753 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -10.32 |
| Energy contribution | -9.95 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.93 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15035405 100 - 23011544 ---UCCGUUCAGUUCGAGUUGAGCGCGCUAACUGCAAGAGCUUAGUUUGGA-AUCGGGCUUAAUAUGCUGGCUUGGAUCCCAAACAAGCGGAGCUUUAAGCUCA----- ---..(((((((......))))))).((.....))..(((((((((((((.-((((((((.........))))).))).))))))((((...))))))))))).----- ( -32.50, z-score = -0.85, R) >droGri2.scaffold_15252 860083 94 - 17193109 ---UUCGUUCCGUUCGAGCUGAGCUCGUCUACUGCUUGACUUGAGCUUUGU-UCGAAGCUUAAUAUGCUGA-CGAAAGCU--AAAAGCUUU-GA--AAAGCUUU----- ---((((((....((((((.(((((((((........)))..)))))).))-))))(((.......)))))-))))....--.(((((((.-..--.)))))))----- ( -30.80, z-score = -2.56, R) >droMoj3.scaffold_6500 23860843 101 + 32352404 ACCACCGUAACGUUUGAGCUGAGCGCGCUUACUGUAUUGCUUGAGCGCUGU-UCCGAGCUUAAUAUGCUGAGCGAAAACUGAAAAAGCUUUCGC--AAAGCUCU----- .............((((((((((((((((((..(.....).))))))).))-))..)))))))...(((..((((((.((.....)).))))))--..)))...----- ( -32.50, z-score = -2.05, R) >droVir3.scaffold_12963 15622099 97 + 20206255 GCUUCCGUUGCGUUCGAGCUGAGCGCGCCUACUGCAAUACUUGAGCUCCGU-UCAGAGCUUAAUAUGCUGG-CUAAAGCU--UAAAGCUUU-GC--AAAGCUCU----- ((.......))....(((((..((..(((....(((.((.((((((((...-...))))))))))))).))-).(((((.--....)))))-))--..))))).----- ( -32.50, z-score = -1.19, R) >droWil1.scaffold_180764 3584710 106 - 3949147 ---ACUGUUUACUUUGAGCCAAGCGCGUUAACUGACCGAUCUUGAUGCAGAUGUCGAGCUUAAUAUGUUUUUCAGAGAAGCUUUUUAUUAAAAACAAAAGCUUUGCUUA ---...........(((((.(((((((((((..........))))))).((((..((((((....((.....))...))))))..))))..........)))).))))) ( -23.30, z-score = -0.83, R) >droPer1.super_5 2613825 97 - 6813705 ---ACCGUUCGAUACGAGCUUUGGGCGCUGAGUGCAAGAUCUGUGCUCGAA-GACGAGCAUAAUAUACU--CUGGCAGACCGAUCGCGCA-AGCUUAAAGCUCA----- ---............(((((((((((.....((((..((((((((((((..-..)))))))).......--..((....)))))))))).-.))))))))))).----- ( -36.70, z-score = -2.50, R) >dp4.chr4_group1 2686667 97 + 5278887 ---ACCGUUCGAUACGAGCUUUGGGCGCUGAGUGCAAGAUCUGUGCUCGAA-GACGAGCGUAAUAUACU--CUGGCAGACCGAUCGUCCA-AGCUUAAAGCUCA----- ---............(((((((((((((((((((((.....)))(((((..-..))))).......)))--).))).(((.....)))..-.))))))))))).----- ( -34.00, z-score = -1.83, R) >droAna3.scaffold_12916 15354927 99 + 16180835 ---UCCGUUCGAUUCGAGCUGAGCCCGCUCACUGCAAGAGAUUGGCCUGGA-GAC-GGCUUAAUAUGUUGGCUUGGCUCCACUUGGAGCAGAGCUUAAAGCUUG----- ---...((((.(...((((..((((.((.....))..((.((.((((.(..-..)-))))....)).))))))..))))....).)))).((((.....)))).----- ( -34.60, z-score = -1.06, R) >droEre2.scaffold_4929 6775231 100 - 26641161 ---UCCGUUCAGUUCGAGUUGAGUGCGCUAACUGCAAGAGCUUAGACCGGA-GUCGGGCUUAAUAUGCUGGCUUGGAUCCACAACAAGCUAAGCUUUAAGCUCA----- ---((((....(((..((((...((((.....))))..))))..)))))))-...((((((((...(((((((((.........)))))).))).)))))))).----- ( -34.40, z-score = -1.68, R) >droYak2.chr2L 2011635 100 + 22324452 ---UCCGUUCACUUUCAGUUGAGCCCGCUAACUGCAAGAGCUUAGAUCGGA-GUCAGGCUUAAUAUGCUGGCUUGGAUCCAAAACAAGCUGAGCUUUAAGCUCA----- ---...(((((........)))))..((.....))..(((((((((((.((-(((((.(.......)))))))).))))......((((...))))))))))).----- ( -30.90, z-score = -1.18, R) >droSec1.super_3 1297524 100 - 7220098 ---UCCGUUCAGUUCGAGUUCAGUGCGCUAACUGCAAGAGCUUAGUAUGGA-GUCGGGCUUAAUAUGCUGGCUUGGAUCUCGAACAAGCAGAGCUUUAAGCUCA----- ---(((((..(((((.....((((......))))...)))))....)))))-...((((((((...(((.(((((.........)))))..))).)))))))).----- ( -33.50, z-score = -1.30, R) >droSim1.chr2L 14782436 100 - 22036055 ---UCCGUUCAGUUCGAGUUCAGUGCGCUAACUGCAAGAGCUUAGUUUGGA-GUCGGGCUUAAUAUGCUGGCUUGGAUCCCAAACAAGCAGAGCUUUAAGCUCA----- ---((((..((((((.....((((......))))...)))))..)..))))-...((((((((...(((.(((((.........)))))..))).)))))))).----- ( -32.50, z-score = -1.22, R) >consensus ___UCCGUUCAGUUCGAGCUGAGCGCGCUAACUGCAAGAGCUUAGCUCGGA_GUCGAGCUUAAUAUGCUGGCUUGGAUCCCAAACAAGCAGAGCUUAAAGCUCA_____ ...............(((((....((((.....))........((((((.....))))))................................))....)))))...... (-10.32 = -9.95 + -0.37)
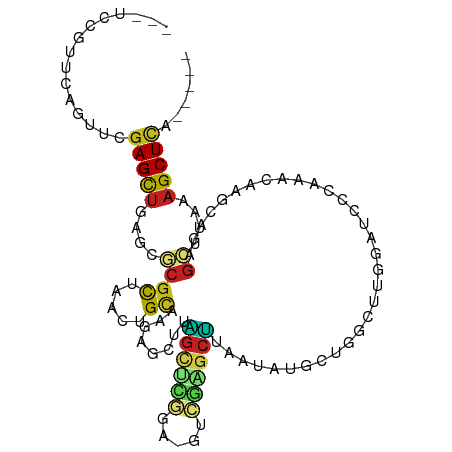

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:42:00 2011