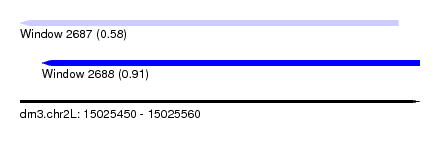
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,025,450 – 15,025,560 |
| Length | 110 |
| Max. P | 0.906234 |

| Location | 15,025,450 – 15,025,554 |
|---|---|
| Length | 104 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.66 |
| Shannon entropy | 0.57656 |
| G+C content | 0.49031 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -13.88 |
| Energy contribution | -13.64 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.575921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
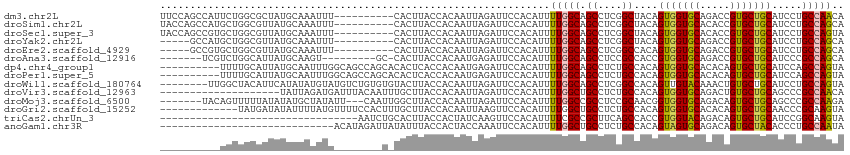
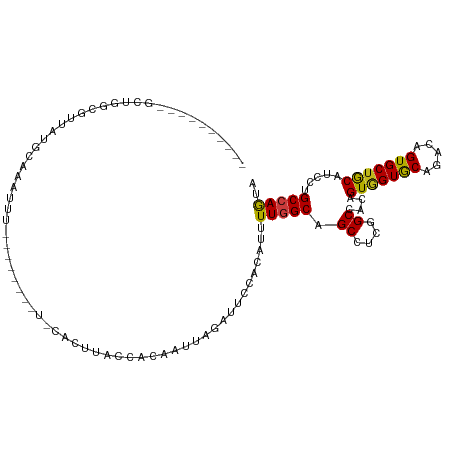
>dm3.chr2L 15025450 104 - 23011544 ---CCAUUCUGGCGCUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCUACAGUGGUGCAGACCGUGCUGCAUCCUGCCAACAGCAUCA ---......(((((..(((((.....----------.....(((((.........(((.....))).(((....)))...)))))(((.....))))))))..)))))......... ( -25.50, z-score = -0.14, R) >droSim1.chr2L 14769304 104 - 22036055 ---CCAUGCUGGCGUUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCUACAGUGGUGCACACCGUGCUGCAUCCUGCCAGCAGCAUCA ---...((((((((..(((((.....----------.....(((((.........(((.....))).(((....)))...)))))((((...)))))))))..))))))))...... ( -35.20, z-score = -2.50, R) >droSec1.super_3 1287368 104 - 7220098 ---CCGUGCUGGCGUUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCUACAGUGGUGCACACCGUGCUGCAUCCUGCCAGUAGCAUCA ---...((((((((..(((((.....----------.....(((((.........(((.....))).(((....)))...)))))((((...)))))))))..))))))))...... ( -33.10, z-score = -1.77, R) >droYak2.chr2L 15134003 104 - 22324452 ---CCAUGCUGGCGUUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCUACAGUGGUGCAGACCGUGCUGCAUCCUGCCAGCAGCAUCA ---...((((((((..(((((.....----------.....(((((.........(((.....))).(((....)))...)))))(((.....))))))))..))))))))...... ( -33.30, z-score = -1.63, R) >droEre2.scaffold_4929 6765164 104 - 26641161 ---CCGUGCUGGCGUUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCCACAGUGGUGCAGACCGUGCUGCAUCCUGCCAGCAGCAUCA ---...((((((((..(((((.....----------.....(((((.....(....)......((((.......))))..)))))(((.....))))))))..))))))))...... ( -31.70, z-score = -0.82, R) >droAna3.scaffold_12916 15345032 103 + 16180835 ----UCGUCUGGCAUUAUGCAAGU---------GC-CACUUACCACAAUGAGAUUCCACAUUUUGGCAGCCUCCGCCACCGUGGUGCAGACCGUGCUGCAUCCCGCCAGCAACAUGA ----..(.(((((...(((((.(.---------.(-..((((((((((((........)))).((((.......))))..)))))).))...)..))))))...))))))....... ( -31.30, z-score = -1.16, R) >dp4.chr4_group1 2675828 110 + 5278887 -------UUUUGCAUUAUGCAAUUUGGCAGCCAGCACACUCACCACAAUGAGAUUCCACAUUUUGGCAGCCUCUGCCACAGUGGUGCACACAGUGCUGCAUCCAGCCAGUAGCAUGA -------.......((((((...(((((...((((((...((((((((((........)))).((((((...))))))..))))))......))))))......)))))..)))))) ( -36.10, z-score = -1.55, R) >droPer1.super_5 2603368 110 - 6813705 -------UUUUGCAUUAUGCAAUUUGGCAGCCAGCACACUCACCACAAUGAGAUUCCACAUUUUGGCAGCCUCUGCCACAGUGGUGCACACAGUGCUGCAUCCAGCCAGUAGCAUGA -------.......((((((...(((((...((((((...((((((((((........)))).((((((...))))))..))))))......))))))......)))))..)))))) ( -36.10, z-score = -1.55, R) >droWil1.scaffold_180764 3568939 112 - 3949147 UUGGCUACAUUCAUAUAUGUAUGUCU-----GUGUGUACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCCACAGUUGUACAAACUGUGCUGCAUCCUGCCAGUAACAUGA ..((((((((......))))).)))(-----(((.((....))))))...........(((.(((((((...((((.((((((.....))))))))))....)))))))....))). ( -31.20, z-score = -1.32, R) >droVir3.scaffold_12963 15612965 97 + 20206255 -------------UAGAUG--AUUUA-----CAAUUUGCUUACCACAAUUAGAUUCCACAUUUUGGCUGCCUCUGCCACAGUGGUGCAGACUGUGCUGCAGCCCGCCAACAGCAUCA -------------..((((--.....-----...(((((..(((((.....(....)......((((.......))))..))))))))))((((...((.....))..)))))))). ( -23.30, z-score = -0.24, R) >droMoj3.scaffold_6500 23852147 104 + 32352404 --------UUUUAUAUAUGCUAUAUU-----CAAUUGGCUUACCACAAUUAGAUUCCACAUUUUGGCCGCCUCCGCAACGGUGGUGCAGACAGUGCUGCAGCCCGCCAAGAGCAUCA --------........((((.....(-----((((((........))))).)).......(((((((.((....))...((.(.(((((......))))).)))))))))))))).. ( -26.80, z-score = -0.74, R) >droGri2.scaffold_15252 851190 101 - 17193109 ---------AUAUUUUAUG--UUUUC-----CACUUUGCUUACCACAAUUAAGUUCCACAUUUUGGCUGCCUCUGCCACAGUGGUGCACACAGUGCUGCAACCCGCAAGUAGCAUCA ---------........((--(...(-----((((..(((((.......))))).........((((.......)))).))))).)))....(((((((.........))))))).. ( -21.40, z-score = -0.16, R) >triCas2.chrUn_3 19677 87 + 565036 -------------------------A-----AUCUGCACUUACCACUAUCAAGUUCCACAUUUUCGCCGCUUCAGCCACCGUGGUACAGACAGUGCUGCAUCCGGCAAGUAACAUCA -------------------------.-----......((((...(((....)))...........(((((....))....(..((((.....))))..)....)))))))....... ( -15.40, z-score = -0.10, R) >anoGam1.chr3R 568652 91 + 53272125 ----------------ACAUAGAUUA----------UAUUUACCACUACCAAAUUCCACAUUUUGGCUGCCUCUGCCACAGUAGUGCAGACAGUGCUACACCCUGCCAAUAGCAUGA ----------------..........----------............(((((........)))))((((..((((....)))).))))...((((((...........)))))).. ( -16.60, z-score = -0.46, R) >consensus ________CUGGCGUUAUGCAAAUUU__________CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCCACAGUGGUGCAGACAGUGCUGCAUCCUGCCAGUAGCAUCA ..............................................................(((((.((....))....(((((((.....))))))).....)))))........ (-13.88 = -13.64 + -0.24)
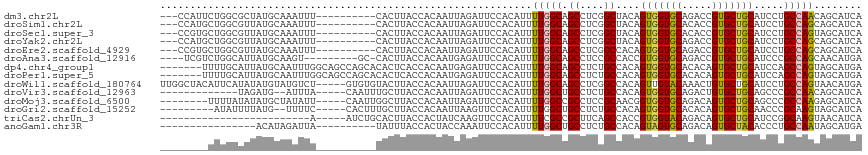
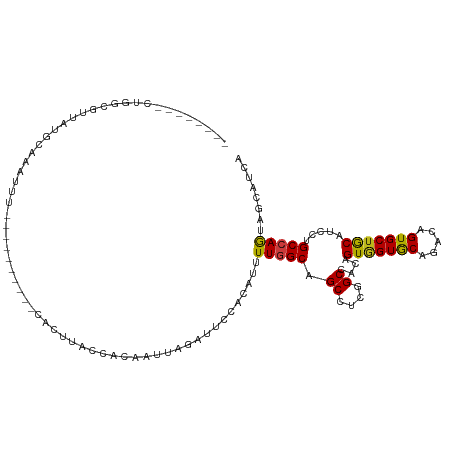
| Location | 15,025,456 – 15,025,560 |
|---|---|
| Length | 104 |
| Sequences | 14 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.92 |
| Shannon entropy | 0.68509 |
| G+C content | 0.48978 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -13.88 |
| Energy contribution | -13.64 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15025456 104 - 23011544 UUCCAGCCAUUCUGGCGCUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCUACAGUGGUGCAGACCGUGCUGCAUCCUGCCAACA .....(((.....))).............----------..........................((((((((....)))..((.(((((((......)))))))))))))).. ( -25.70, z-score = -0.31, R) >droSim1.chr2L 14769310 104 - 22036055 UACCAGCCAUGCUGGCGUUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCUACAGUGGUGCACACCGUGCUGCAUCCUGCCAGCA .........((((((((..(((((.....----------.....(((((.........(((.....))).(((....)))...)))))((((...)))))))))..)))))))) ( -34.40, z-score = -2.27, R) >droSec1.super_3 1287374 104 - 7220098 UACCAGCCGUGCUGGCGUUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCUACAGUGGUGCACACCGUGCUGCAUCCUGCCAGUA .........((((((((..(((((.....----------.....(((((.........(((.....))).(((....)))...)))))((((...)))))))))..)))))))) ( -32.30, z-score = -1.47, R) >droYak2.chr2L 15134009 99 - 22324452 -----GCCAUGCUGGCGUUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCUACAGUGGUGCAGACCGUGCUGCAUCCUGCCAGCA -----....((((((((..(((((.....----------.....(((((.........(((.....))).(((....)))...)))))(((.....))))))))..)))))))) ( -32.50, z-score = -1.62, R) >droEre2.scaffold_4929 6765170 99 - 26641161 -----GCCGUGCUGGCGUUAUGCAAAUUU----------CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCCACAGUGGUGCAGACCGUGCUGCAUCCUGCCAGCA -----....((((((((..(((((.....----------.....(((((.....(....)......((((.......))))..)))))(((.....))))))))..)))))))) ( -30.90, z-score = -0.81, R) >droAna3.scaffold_12916 15345038 97 + 16180835 -------UCGUCUGGCAUUAUGCAAGU---------GC-CACUUACCACAAUGAGAUUCCACAUUUUGGCAGCCUCCGCCACCGUGGUGCAGACCGUGCUGCAUCCCGCCAGCA -------..(.(((((...(((((.(.---------.(-..((((((((((((........)))).((((.......))))..)))))).))...)..))))))...)))))). ( -31.30, z-score = -1.55, R) >dp4.chr4_group1 2675834 104 + 5278887 ----------UUUUGCAUUAUGCAAUUUGGCAGCCAGCACACUCACCACAAUGAGAUUCCACAUUUUGGCAGCCUCUGCCACAGUGGUGCACACAGUGCUGCAUCCAGCCAGUA ----------..((((.....)))).(((((...((((((...((((((((((........)))).((((((...))))))..))))))......))))))......))))).. ( -32.90, z-score = -1.52, R) >droPer1.super_5 2603374 104 - 6813705 ----------UUUUGCAUUAUGCAAUUUGGCAGCCAGCACACUCACCACAAUGAGAUUCCACAUUUUGGCAGCCUCUGCCACAGUGGUGCACACAGUGCUGCAUCCAGCCAGUA ----------..((((.....)))).(((((...((((((...((((((((((........)))).((((((...))))))..))))))......))))))......))))).. ( -32.90, z-score = -1.52, R) >droWil1.scaffold_180764 3568945 106 - 3949147 --------UUGGCUACAUUCAUAUAUGUAUGUCUGUGUGUACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCCACAGUUGUACAAACUGUGCUGCAUCCUGCCAGUA --------..((((((((......))))).)))((((.((....))))))...............(((((((...((((.((((((.....))))))))))....))))))).. ( -30.70, z-score = -1.71, R) >droVir3.scaffold_12963 15612971 94 + 20206255 --------------------UAUUAGAUGAUUUACAAUUUGCUUACCACAAUUAGAUUCCACAUUUUGGCUGCCUCUGCCACAGUGGUGCAGACUGUGCUGCAGCCCGCCAACA --------------------.......((...((((.(((((..(((((.....(....)......((((.......))))..)))))))))).))))...))........... ( -18.80, z-score = 0.58, R) >droMoj3.scaffold_6500 23852153 104 + 32352404 -------UACAGUUUUUAUAUAUGCUAUAUU---CAAUUGGCUUACCACAAUUAGAUUCCACAUUUUGGCCGCCUCCGCAACGGUGGUGCAGACAGUGCUGCAGCCCGCCAAGA -------................((((....---....)))).....................(((((((.((....))...((.(.(((((......))))).)))))))))) ( -23.20, z-score = 0.22, R) >droGri2.scaffold_15252 851196 101 - 17193109 -------------UAUGAUAUAUUUUAUGUUUUCCACUUUGCUUACCACAAUUAAGUUCCACAUUUUGGCUGCCUCUGCCACAGUGGUGCACACAGUGCUGCAACCCGCAAGUA -------------(((((......))))).....((((.((..((((((......(.....)....((((.......))))..))))))..)).)))).(((.....))).... ( -19.30, z-score = 0.38, R) >triCas2.chrUn_3 19683 81 + 565036 ---------------------------------AAUCUGCACUUACCACUAUCAAGUUCCACAUUUUCGCCGCUUCAGCCACCGUGGUACAGACAGUGCUGCAUCCGGCAAGUA ---------------------------------.......((((...(((....)))...........(((((....))....(..((((.....))))..)....))))))). ( -15.40, z-score = -0.05, R) >anoGam1.chr3R 568658 85 + 53272125 -----------------------------ACAUAGAUUAUAUUUACCACUACCAAAUUCCACAUUUUGGCUGCCUCUGCCACAGUAGUGCAGACAGUGCUACACCCUGCCAAUA -----------------------------.................((((.(((((........)))))((((..((((....)))).))))..))))................ ( -15.30, z-score = -0.95, R) >consensus __________GCUGGCGUUAUGCAAAUUU________U_CACUUACCACAAUUAGAUUCCACAUUUUGGCAGCCUCGGCCACAGUGGUGCAGACAGUGCUGCAUCCUGCCAGUA .................................................................(((((.((....))....(((((((.....))))))).....))))).. (-13.88 = -13.64 + -0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:58 2011