| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,997,615 – 14,997,721 |
| Length | 106 |
| Max. P | 0.603683 |

| Location | 14,997,615 – 14,997,721 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.12 |
| Shannon entropy | 0.73340 |
| G+C content | 0.42231 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -12.24 |
| Energy contribution | -11.97 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.34 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
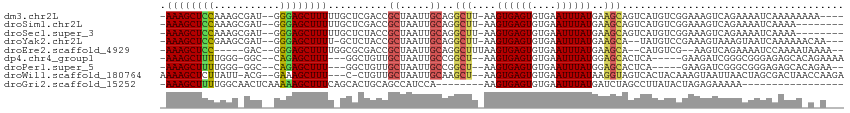
>dm3.chr2L 14997615 106 + 23011544 -AAAGCUCCAAAGCGAU--GGGAGCUUUUUGCUCGACCGCUAAUUGCAGGCUU-AAGUGAGUGUGAAUUUAUGAAGCAGUCAUGUCGGAAAGUCAGAAAAUCAAAAAAAA---- -...(((((........--.)))))((((((((((((.((.....))..((((-..((((((....)))))).))))......))))...)).))))))...........---- ( -23.20, z-score = 0.18, R) >droSim1.chr2L 14741386 102 + 22036055 -AAAGCUCCAAAGCGAU--GGGAGCUUUUUGCUCGACCGCUAAUUGCAGGCUU-AAGUGAGUGUGAAUUUAUGAAGCAGUCAUGUCGGAAAGUCAGAAAAUCAAAA-------- -...(((((........--.)))))((((((((((((.((.....))..((((-..((((((....)))))).))))......))))...)).)))))).......-------- ( -23.20, z-score = 0.30, R) >droSec1.super_3 1259869 102 + 7220098 -AAAGCUCCAAAGCGAU--GGGAGCUUUUUGCUCUACCGCUAAUUGCAGGCUU-AAGUGAGUGUGAAUUUAUGAAGCAGUCAUGUCGGAAAGUCAGAAAAUCAAAA-------- -...((.....((((..--.(((((.....)))))..))))....)).(((((-..((((.(((...........))).))))......)))))............-------- ( -24.20, z-score = -0.16, R) >droYak2.chr2L 15103630 104 + 22324452 -AAAGCUCCGAAGCGAU--GGGAGCUUUU-GCUCUACCGCUAAUUGCAGGCUU-AAGUGAGUGUGAAUUUAUGAAGCA--UAUGUCCGAAAGUAAAGUAAUCAAAAAACAA--- -.(((((((.(.....)--.)))))))((-(((.(((.((.....))..((((-..((((((....)))))).)))).--...........))).)))))...........--- ( -23.00, z-score = -0.45, R) >droEre2.scaffold_4929 6735359 100 + 26641161 -AAAGCUCC-----GAC--GGGAGCUUUUGGCGCGACCGCUAAUUGCAGGCUUUAAGUGAGUGUGAAUUUAUGAAGCA--CAUGUCG--AAGUCAGAAAAUCCAAAAUAAAA-- -...(((((-----...--.)))))(((((((.((((.((.....))..(((((..((((((....))))))))))).--...))))--..)))))))..............-- ( -28.30, z-score = -1.76, R) >dp4.chr4_group1 2635084 100 - 5278887 -AAAGCUUUUGGG-GGC--CAGAGCUUU---GGCUGUUGCUAAUUGCCGGCU--AAGUGAGUGUGAAUUUAUGGAGCACUCA-----GAAGAUCGGGCGGGAGAGCACAGAAAA -((((((((.((.-..)--)))))))))---..((((.(((..(((((.(..--...(((((((...........)))))))-----......).)))))...))))))).... ( -31.20, z-score = -0.82, R) >droPer1.super_5 2562835 98 + 6813705 -AAAGCUUUUGGG-GGC--CAGAGCUUU---GGCUGUUGCUAAUUGCCGGCU--AAGUGAGUGUGAAUUUAUGGAGCACUCA-----GAAGAUCGGGCGGGAGAGCACAGAA-- -((((((((.((.-..)--)))))))))---..((((.(((..(((((.(..--...(((((((...........)))))))-----......).)))))...)))))))..-- ( -31.20, z-score = -0.76, R) >droWil1.scaffold_180764 3518300 105 + 3949147 AAAAGCUCUUAUU-ACG--GAAAGCUUU---C-CUGUUGCUAAUUGCAAGCU--AAGUGAGUGUGAAUUUAUAAGGUAGUCACUACAAAGUAAUUAACUAGCGACUAACCAAGA ....((.((((((-..(--(((....))---)-)..((((.....))))...--.)))))).))..........(((((((.(((..((....))...))).)))).))).... ( -21.30, z-score = -0.19, R) >droGri2.scaffold_15252 822211 88 + 17193109 -AAAGCUUUUGGCAACUCAAAAAGCUUUCAGCACUGCAGCCAUCCA--------AAGUGAGUGUGAAUUUAUGAUCUAGCCUUAUACUAGAGAAAAA----------------- -((((((((((....)...)))))))))..(((((...((......--------..)).)))))..........(((((.......)))))......----------------- ( -15.30, z-score = 0.61, R) >consensus _AAAGCUCCUAAG_GAU__GGGAGCUUUU_GCUCUACCGCUAAUUGCAGGCUU_AAGUGAGUGUGAAUUUAUGAAGCAGUCAUGUCGGAAAGUCAGAAAAUCAAAAA_A_A___ .((((((((...........))))))))..........((.....))..(((....((((((....))))))..)))..................................... (-12.24 = -11.97 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:57 2011