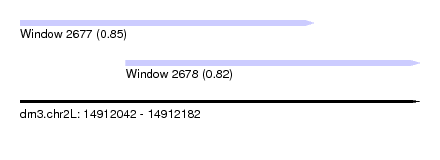
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,912,042 – 14,912,182 |
| Length | 140 |
| Max. P | 0.851486 |

| Location | 14,912,042 – 14,912,145 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.46 |
| Shannon entropy | 0.55902 |
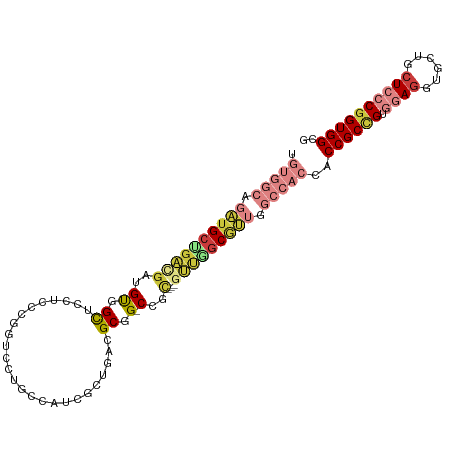
| G+C content | 0.68313 |
| Mean single sequence MFE | -48.97 |
| Consensus MFE | -23.90 |
| Energy contribution | -27.46 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
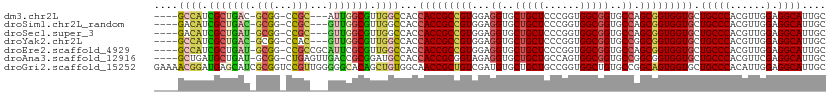
>dm3.chr2L 14912042 103 + 23011544 UGUGGCAGAUGCUGACGAUGUGGCUCCUCCCGGUCCAGCCAUCGCUGACGCGG-CCGC---AUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCG .(((((.(((((...((((((((((((....))..((((....))))....))-))))---))))))))).)))))..((((((.((((......)))))))))).. ( -50.10, z-score = -1.06, R) >droSim1.chr2L_random 572143 103 + 909653 UGUGGCAGAUGCUGACGAUGUAGCUCCUCCCGGUCCUGACAUCGCUGACGCGG-CCGC---GUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCG .(((((.(((((..(((((((((..((....))..)).)))))((((...)))-)...---))..))))).)))))..((((((.((((......)))))))))).. ( -49.00, z-score = -1.40, R) >droSec1.super_3 1182099 103 + 7220098 UGUGGCAGAUGCUGACGAUGUAGCUCCUCCCGGACCUGACAUCGCUGAUGCGG-CCGC---GUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCG .(((((.(((((..(((((((((.(((....))).)).)))))((((...)))-)...---))..))))).)))))..((((((.((((......)))))))))).. ( -50.50, z-score = -2.25, R) >droYak2.chr2L 15024221 103 + 22324452 CGUGGCGGAUGCCGACGAUGUGGCUCCUCCCGGUCCUGCCAUCGCUGACGCGG-CCAC---GUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCG .(((((.((((((((((..(((((.((....))....)))))(((....))).-...)---))))))))).)))))..((((((.((((......)))))))))).. ( -55.50, z-score = -2.20, R) >droEre2.scaffold_4929 6657583 106 + 26641161 UGUGGCAGAUGCGGACGAUGUGGCGCCUCCUGGUCCUGCCAUCGCUGAUGCGG-CCGCCGCAUUCGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCG .(((((.((((((...(((((((((((...(((.....)))..((....))))-.))))))))))))))).)))))..((((((.((((......)))))))))).. ( -55.30, z-score = -1.87, R) >droAna3.scaffold_12916 15243130 100 - 16180835 ------UGCGGCUGACACUGCCGUCCCUGCUGCGGAUGCUGAUGCUGAUGCGG-CUGAGUUGACCGCGGAUGCCACCACCGCGGUAGAGGUGCUGCUGCCAGUGGCG ------.((.((((.((..((.(((((((((((((.((.((..((...(((((-(......).)))))...)))).))))))))))).)).)).)))).)))).)). ( -43.30, z-score = -1.08, R) >droGri2.scaffold_15252 706161 97 + 17193109 ---------UGCUGUUGCUGUUGCAACAGUGGAAAACGG-AUGAGCAUCGCGGUCCGUUGGGGGCACAGCUGUGGCAACCGCUGUCGAUGUGCUGCUGCCGGUGGCU ---------.((((((((....))))))))....(((((-((..((...)).)))))))((.((((((.(.(..((....))..).).)))))).))(((...))). ( -39.10, z-score = -0.34, R) >consensus UGUGGCAGAUGCUGACGAUGUGGCUCCUCCCGGUCCUGCCAUCGCUGACGCGG_CCGC___GUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCG .(((((.(((((((((((((.....((....))......))))((....))..........))))))))).)))))..((((((.((((......)))))))))).. (-23.90 = -27.46 + 3.56)


| Location | 14,912,079 – 14,912,182 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.02 |
| Shannon entropy | 0.32866 |
| G+C content | 0.70184 |
| Mean single sequence MFE | -56.34 |
| Consensus MFE | -41.22 |
| Energy contribution | -41.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14912079 103 + 23011544 ----GCCAUCGCUGAC-GCGG-CCGC---AUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCGGUGCCAGCGGUGGUGCUGCCCACGUUGGAGGCAUUGC ----(((....(..((-(.((-(.((---((((.(((((((.(((.((((((.((((......)))))))))).)))))))))).)))))).)))..)))..).)))..... ( -61.20, z-score = -2.65, R) >droSim1.chr2L_random 572180 103 + 909653 ----GACAUCGCUGAC-GCGG-CCGC---GUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCGGUGCCAGCGGUGGUGCUGCCCACGUUGGAGGCAUUGC ----..((((((..((-((..-..))---))..))((((((.(((.((((((.((((......)))))))))).)))))))))))))(((((..((.....)).)))))... ( -58.90, z-score = -2.20, R) >droSec1.super_3 1182136 103 + 7220098 ----GACAUCGCUGAU-GCGG-CCGC---GUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCGGUGCCAGCGGUGGUGCUGCCCACGUUGGAGGCAUUGC ----..((((((..((-((..-..))---))..))((((((.(((.((((((.((((......)))))))))).)))))))))))))(((((..((.....)).)))))... ( -57.10, z-score = -1.95, R) >droYak2.chr2L 15024258 103 + 22324452 ----GCCAUCGCUGAC-GCGG-CCAC---GUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCGGUGCCGGCGGUGGUGCUGCCCACGUUGGAGGCAUUGC ----((((((((((..-((((-((((---(....)).)))))(((.((((((.((((......)))))))))).)))))))))))))))(((..((.....)).)))..... ( -57.40, z-score = -1.38, R) >droEre2.scaffold_4929 6657620 106 + 26641161 ----GCCAUCGCUGAU-GCGG-CCGCCGCAUUCGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCGGUGCCAGCGGUGGUGCUGCCCACGUUGGAGGCAUUGC ----((((((((((..-((((-(((.(((....))).)))))(((.((((((.((((......)))))))))).)))))))))))))))(((..((.....)).)))..... ( -60.60, z-score = -2.35, R) >droAna3.scaffold_12916 15243161 106 - 16180835 ----GCUGAUGCUGAU-GCGG-CUGAGUUGACCGCGGAUGCCACCACCGCGGUAGAGGUGCUGCUGCCAGUGGCGGUGCCGGCGGUGGUGCUGCCCACGUUCGAGGCAUUGC ----((.((((((...-((((-(......).))))((..(.(((((((((......((..((((((....))))))..)).))))))))).)..))........)))))))) ( -48.00, z-score = -0.23, R) >droGri2.scaffold_15252 706183 112 + 17193109 GAAAACGGAUGAGCAUCGCGGUCCGUUGGGGGCACAGCUGUGGCAACCGCUGUCGAUGUGCUGCUGCCGGUGGCUGUGCCGGCAGUGGUGCUGCCCACAUUGGAGGCAUUGC ............(((..((..((((.((.(((((((((.((....)).)))))....(..(..((((((((......))))))))..)..).)))).)).)))).))..))) ( -51.20, z-score = -0.84, R) >consensus ____GCCAUCGCUGAU_GCGG_CCGC___GUUGGCGUUGGCCACCACCGCCGUGGAGGUGCUGCUCCCGGUGGCGGUGCCAGCGGUGGUGCUGCCCACGUUGGAGGCAUUGC ....((((.(((((((.(((...)))...))))))).))))...(((((((((...((..(((((......)))))..)).))))))))).(((((......).)))).... (-41.22 = -41.33 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:50 2011