| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,911,430 – 14,911,550 |
| Length | 120 |
| Max. P | 0.910802 |

| Location | 14,911,430 – 14,911,550 |
|---|---|
| Length | 120 |
| Sequences | 7 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.50004 |
| G+C content | 0.50945 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.99 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
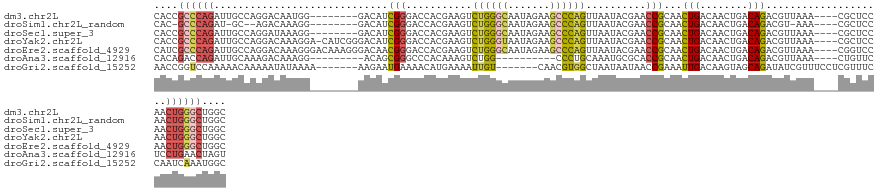
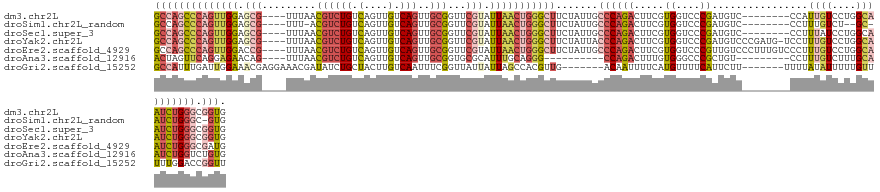
>dm3.chr2L 14911430 120 + 23011544 CACCGCCCAGAUUGCCAGGACAAUGG--------GACAUCGGGACCACGAAGUCUGGGCAAUAGAAGCCCAGUUAAUACGAACCGCAACUGACAACUGACAGACGUUAAA----CGCUCCAACUGGGCUGGC ..(((((((((((.(((......)))--------....(((......))))))))))))......(((((((((....((....((..(((........)))..))....----))....))))))))))). ( -40.70, z-score = -2.95, R) >droSim1.chr2L_random 571474 115 + 909653 CAC-GCCCAGAU-GC--AGACAAAGG--------GACAUCGGGACCACGAAGUCUGGGCAAUAGAAGCCCAGUUAAUACGAACCGCAACUGACAACUGACAGACGU-AAA----CGCUCCAACUGGGCUGGC ...-((((((((-..--.......((--------..(....)..)).....))))))))......(((((((((....((....((..(((........)))..))-...----))....)))))))))... ( -34.74, z-score = -2.05, R) >droSec1.super_3 1181420 120 + 7220098 CACCGCCCAGAUUGCCAGGAUAAAGG--------GACAUCGGGACCACGAAGUCUGGGCAAUAGAAGCCCAGUUAAUACGAACCGCAACUGACAACUGACAGACGUUAAA----CGCUCCAACUGGGCUGGC ..(((((((((((.((........))--------....(((......))))))))))))......(((((((((....((....((..(((........)))..))....----))....))))))))))). ( -38.80, z-score = -2.53, R) >droYak2.chr2L 15023447 127 + 22324452 CACCGCCCAGAUUGCCAGGACAAAGGA-CAUCGGGACAUCGGGACCACGAAGUCUGGGUAAUAGAAGCCCAGUUAAUACGAACCGCAACUGACAACUGACAGACGUUAAA----CGCUCCAACUGGGCUGGC ..(((((((((((.((........)).-....((..(....)..))....)))))))))......(((((((((....((....((..(((........)))..))....----))....))))))))))). ( -37.50, z-score = -1.34, R) >droEre2.scaffold_4929 6656875 128 + 26641161 CAUCGCCCAGAUUGCCAGGACAAAGGGACAAAGGGACAACGGGACCACGAAGUCUGGGCAAUAGAAGCCCAGUUAAUACGAACCGCAACUGACAACUGACAGACGUUAAA----CGGUCCAACUGGGCUGGC ....(((((((((.((........))......((..(....)..))....)))))))))......(((((((((....((....((..(((........)))..))....----))....)))))))))... ( -39.80, z-score = -2.06, R) >droAna3.scaffold_12916 15242576 109 - 16180835 CACAGACCAGAUUGCAAAGACAAAGG---------ACAGCGGGCCCACAAAGUCUGG----------CCCUGCAAAUGCGCACCGCAACUGACAACUGACAGACGUUAAA----CUGUUCUCCUGAACUAGU ..(((..(((.((((...........---------.....(((((..........))----------)))(((......)))..)))))))....)))...........(----((((((....)))).))) ( -24.30, z-score = 0.26, R) >droGri2.scaffold_15252 705520 118 + 17193109 AACCGGUCCAAAAACAAAAAUAUAAAA-------AAGAAUGAAAACAUGAAAAUUGU-------CAACGUGGCUAAUAAUAACCGAAAUUGACAAGUAGCAGAUAUCGUUUCCUCGUUUCCAAUCAAAUGGC ...........................-------..((((((.....((....((((-------(((..(((.((....)).)))...)))))))....))....))))))..((((((......)))))). ( -13.90, z-score = 0.57, R) >consensus CACCGCCCAGAUUGCCAGGACAAAGG________GACAUCGGGACCACGAAGUCUGGGCAAUAGAAGCCCAGUUAAUACGAACCGCAACUGACAACUGACAGACGUUAAA____CGCUCCAACUGGGCUGGC ....((((((.............................(((...........((((((.......))))))..........)))...(((........)))....................)))))).... (-17.35 = -17.99 + 0.63)
| Location | 14,911,430 – 14,911,550 |
|---|---|
| Length | 120 |
| Sequences | 7 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.50004 |
| G+C content | 0.50945 |
| Mean single sequence MFE | -39.21 |
| Consensus MFE | -17.60 |
| Energy contribution | -19.67 |
| Covariance contribution | 2.07 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
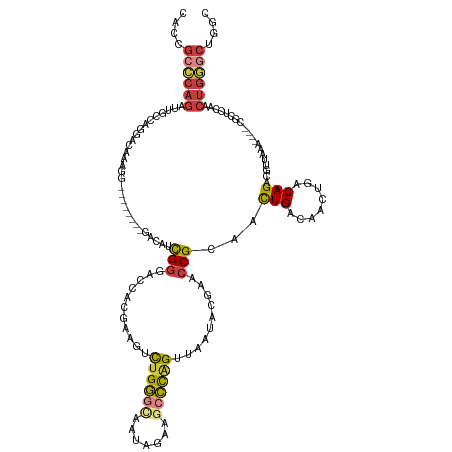
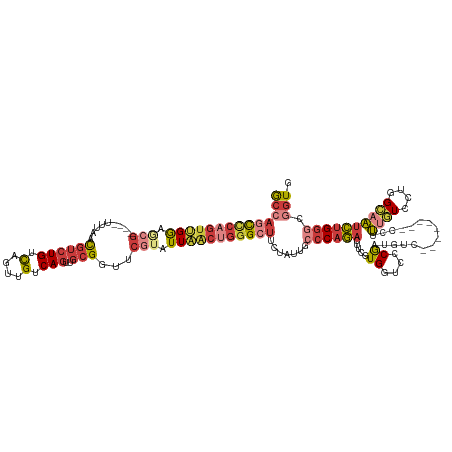
>dm3.chr2L 14911430 120 - 23011544 GCCAGCCCAGUUGGAGCG----UUUAACGUCUGUCAGUUGUCAGUUGCGGUUCGUAUUAACUGGGCUUCUAUUGCCCAGACUUCGUGGUCCCGAUGUC--------CCAUUGUCCUGGCAAUCUGGGCGGUG ((((((((((((((.(((----.....((((((.(....).)))..)))...))).)))))))))))......(((((((....((((..(....)..--------))))(((....))).)))))))))). ( -46.70, z-score = -3.20, R) >droSim1.chr2L_random 571474 115 - 909653 GCCAGCCCAGUUGGAGCG----UUU-ACGUCUGUCAGUUGUCAGUUGCGGUUCGUAUUAACUGGGCUUCUAUUGCCCAGACUUCGUGGUCCCGAUGUC--------CCUUUGUCU--GC-AUCUGGGC-GUG ...(((((((((((.(((----...-.((((((.(....).)))..)))...))).)))))))))))......(((((((((....))))..(((((.--------.........--))-))))))))-... ( -41.40, z-score = -2.74, R) >droSec1.super_3 1181420 120 - 7220098 GCCAGCCCAGUUGGAGCG----UUUAACGUCUGUCAGUUGUCAGUUGCGGUUCGUAUUAACUGGGCUUCUAUUGCCCAGACUUCGUGGUCCCGAUGUC--------CCUUUAUCCUGGCAAUCUGGGCGGUG ((((((((((((((.(((----.....((((((.(....).)))..)))...))).)))))))))))......((((((((.....)(((..((((..--------....))))..)))..)))))))))). ( -45.20, z-score = -3.08, R) >droYak2.chr2L 15023447 127 - 22324452 GCCAGCCCAGUUGGAGCG----UUUAACGUCUGUCAGUUGUCAGUUGCGGUUCGUAUUAACUGGGCUUCUAUUACCCAGACUUCGUGGUCCCGAUGUCCCGAUG-UCCUUUGUCCUGGCAAUCUGGGCGGUG ((((((((((((((.(((----.....((((((.(....).)))..)))...))).))))))))))).......((((((..(((.((.(.....).)))))((-.((........)))).)))))).))). ( -44.20, z-score = -2.13, R) >droEre2.scaffold_4929 6656875 128 - 26641161 GCCAGCCCAGUUGGACCG----UUUAACGUCUGUCAGUUGUCAGUUGCGGUUCGUAUUAACUGGGCUUCUAUUGCCCAGACUUCGUGGUCCCGUUGUCCCUUUGUCCCUUUGUCCUGGCAAUCUGGGCGAUG ...(((((((((((((((----(.......(((.(....).)))..))))))).....)))))))))..(((((((((((......((..(....)..)).........((((....))))))))))))))) ( -44.20, z-score = -3.07, R) >droAna3.scaffold_12916 15242576 109 + 16180835 ACUAGUUCAGGAGAACAG----UUUAACGUCUGUCAGUUGUCAGUUGCGGUGCGCAUUUGCAGGG----------CCAGACUUUGUGGGCCCGCUGU---------CCUUUGUCUUUGCAAUCUGGUCUGUG (((((....(((((((((----........))))(((..(.(((((((((.......)))))(((----------((..........))))))))).---------)..)))))))).....)))))..... ( -27.90, z-score = 1.35, R) >droGri2.scaffold_15252 705520 118 - 17193109 GCCAUUUGAUUGGAAACGAGGAAACGAUAUCUGCUACUUGUCAAUUUCGGUUAUUAUUAGCCACGUUG-------ACAAUUUUCAUGUUUUCAUUCUU-------UUUUAUAUUUUUGUUUUUGGACCGGUU (((....((.((((((((.(....)............((((((((...(((((....)))))..))))-------))))......)))))))).))..-------............(((....))).))). ( -24.90, z-score = -1.74, R) >consensus GCCAGCCCAGUUGGAGCG____UUUAACGUCUGUCAGUUGUCAGUUGCGGUUCGUAUUAACUGGGCUUCUAUUGCCCAGACUUCGUGGUCCCGAUGUC________CCUUUGUCCUGGCAAUCUGGGCGGUG ((((((((((((((.(((.........((((((.(....).)))..)))...))).))))))))))).......((((((.....((....))................((((....)))))))))).))). (-17.60 = -19.67 + 2.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:48 2011