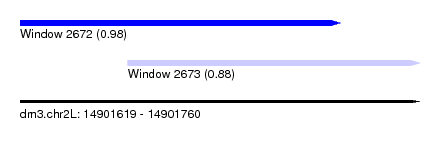
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,901,619 – 14,901,760 |
| Length | 141 |
| Max. P | 0.980982 |

| Location | 14,901,619 – 14,901,732 |
|---|---|
| Length | 113 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.13 |
| Shannon entropy | 0.62490 |
| G+C content | 0.39266 |
| Mean single sequence MFE | -15.93 |
| Consensus MFE | -10.27 |
| Energy contribution | -10.38 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
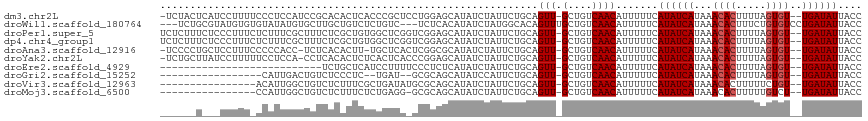
>dm3.chr2L 14901619 113 + 23011544 -UCUACUCAUCCUUUUCCCUCCAUCCGCACACUCACCCGCUCCUGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACC -.................(((((...((..........))...)))))............((....-))((((((((((.....................)))))--)))))..... ( -14.90, z-score = -1.64, R) >droWil1.scaffold_180764 566259 111 - 3949147 ---UCUGCGUAUGUGUGUAUAUGUGCUUGCUGUCUCUGUC---UCUCACAUAUCUAUGGCACAGUUUGCUGUCAACAUUUUUCAUAUCAUAAACACUUUCUGUGUCCUGAUAUUACC ---...(((..(((((..(((((((...((.......)).---...))))))).....)))))...)))..............((((((...((((.....))))..)))))).... ( -20.10, z-score = -0.65, R) >droPer1.super_5 2435740 114 + 6813705 UCUCUUUCUCCCUUUCUCUUUCGCUUUCUCGCUGUGGCUCGGUCGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACC ......................((......)).(..((((((..(((.....)))...))).))).-.)((((((((((.....................)))))--)))))..... ( -18.20, z-score = -1.14, R) >dp4.chr4_group1 2505445 114 - 5278887 UCUCUUUCUCCCUUUCUCUUUCGCUUUCUCGCUGUGGCUCGGUCGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACC ......................((......)).(..((((((..(((.....)))...))).))).-.)((((((((((.....................)))))--)))))..... ( -18.20, z-score = -1.14, R) >droAna3.scaffold_12916 15233035 111 - 16180835 -UCCCCUGCUCCUUUCCCCCACC-UCUCACACUU-UGCUCACUCGGCGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACC -......((..((..........-..........-.(((.....)))(((.........)))))..-))((((((((((.....................)))))--)))))..... ( -12.80, z-score = -1.24, R) >droYak2.chr2L 15004583 112 + 22324452 -UCUGCUUAUCCUUUUUCCUCCA-CCUCACACUCUCACUCACCCGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACC -.((((............((((.-....................))))............))))..-..((((((((((.....................)))))--)))))..... ( -15.35, z-score = -2.20, R) >droEre2.scaffold_4929 6644927 87 + 26641161 ---------------------------UCUGCUCAUCCUUUUCCCUCUCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACC ---------------------------.((((............................))))..-..((((((((((.....................)))))--)))))..... ( -10.49, z-score = -2.12, R) >droGri2.scaffold_15252 16500083 93 - 17193109 -----------------CAUUGACUGUCUCCCUC--UGAU--GCGCAGCAUAUCCAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACC -----------------....((((((.......--..((--((...)))).........))))))-..((((((((((.....................)))))--)))))..... ( -15.87, z-score = -0.77, R) >droVir3.scaffold_12963 4739179 98 + 20206255 ----------------ACAUUGGCUGUCUCUUUCGCUGAUAUGCGCAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUUCUGU--UGAUAUUACC ----------------.....((((((..........(((((((...)))))))......))))))-..((((((((.........................)))--)))))..... ( -15.90, z-score = -0.44, R) >droMoj3.scaffold_6500 8656187 97 + 32352404 ----------------CCAUUGGCUGUCUCUUUCUCUGAGG-GCGCAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUUGUCU--UGAUAUUACC ----------------......((((((((((.....))))-).)))))..............(((-(....)))).......((((((...(((.....)))..--)))))).... ( -17.50, z-score = -0.75, R) >consensus ______U___CCUUU_CCAUUCGCUCUCUCACUCUCUCUCA_CCGGAGCAUAUCUAUUCUGCAGUU_GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU__UGAUAUUACC ............................................................((((....))))...........((((((...((((.....))))..)))))).... (-10.27 = -10.38 + 0.11)

| Location | 14,901,657 – 14,901,760 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Shannon entropy | 0.35335 |
| G+C content | 0.36043 |
| Mean single sequence MFE | -16.11 |
| Consensus MFE | -10.27 |
| Energy contribution | -10.38 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
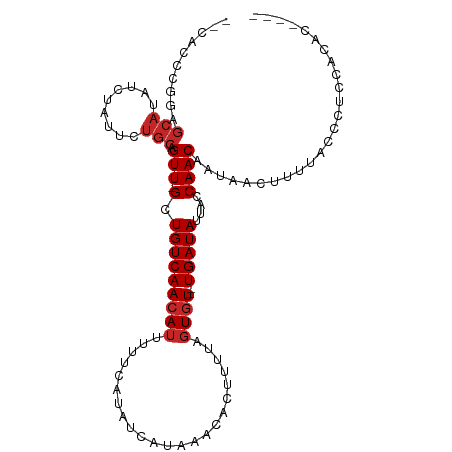
>dm3.chr2L 14901657 103 + 23011544 --CUCCUGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACCAACAAUAACUUUCACGCUCCUCACACAC --.....(((((...........((....-))((((((((((.....................)))))--)))))....................)))))........ ( -18.40, z-score = -2.81, R) >droWil1.scaffold_180764 566299 87 - 3949147 ---------CACAUAUCUAUGGCACAGUUUGCUGUCAACAUUUUUCAUAUCAUAAACACUUUCUGUGUCCUGAUAUUACCAACAAUAACUUUUCUC------------ ---------..........(((.((((....))))...........((((((...((((.....))))..))))))..)))...............------------ ( -12.30, z-score = -1.81, R) >droPer1.super_5 2435779 99 + 6813705 --CGGUCGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACCAACAAUAACUUUUCCCUCACACAC---- --..(..((((((.........))).(((-(.((((((((((.....................)))))--)))))....)))).........)))..)......---- ( -15.40, z-score = -1.43, R) >dp4.chr4_group1 2505484 99 - 5278887 --CGGUCGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACCAACAAUAACUUUUCCCUCACACAC---- --..(..((((((.........))).(((-(.((((((((((.....................)))))--)))))....)))).........)))..)......---- ( -15.40, z-score = -1.43, R) >droAna3.scaffold_12916 15233071 94 - 16180835 --CACUCGGCGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACCAACAAUAACUUUCCCUCGC--------- --.....((.(((.........))).(((-(.((((((((((.....................)))))--)))))....))))..........))....--------- ( -13.00, z-score = -1.22, R) >droSim1.chr2L 14648970 99 + 22036055 --CUCCUGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACCAACAAUAACUUUCACGCUCCUCAC---- --.....(((((...........((....-))((((((((((.....................)))))--)))))....................)))))....---- ( -18.40, z-score = -2.98, R) >droSec1.super_3 1171566 99 + 7220098 --CUCCUGGAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACCAACAAUAACUUUCACGCUCCUCAC---- --.....(((((...........((....-))((((((((((.....................)))))--)))))....................)))))....---- ( -18.40, z-score = -2.98, R) >droGri2.scaffold_15252 16500101 93 - 17193109 --GAUGCGCAGCAUAUCCAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU--UGAUAUUACCAACAAUAACUUUUACCCA---------- --.....((((.((....)).)))).(((-(.((((((((((.....................)))))--)))))....))))...............---------- ( -15.00, z-score = -1.65, R) >droVir3.scaffold_12963 4739200 105 + 20206255 GAUAUGCGCAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUUCUGU--UGAUAUUACCAACAAUAACUUUUACCCGACACAUACGC ((((((.((((.((....)).)))).(((-(....))))......))))))..............(((--((.......)))))........................ ( -15.40, z-score = -1.43, R) >droMoj3.scaffold_6500 8656208 103 + 32352404 -GAGGGCGCAGCAUAUCUAUUCUGCAGUU-GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUUGUCU--UGAUAUUACCAACAAUAACUUUUGCCCA-CACCCACAA -..((((((((((...((.......)).)-)))))...........((((((...(((.....)))..--)))))).................)))).-......... ( -19.40, z-score = -2.26, R) >consensus __CACCCGGAGCAUAUCUAUUCUGCAGUU_GCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGU__UGAUAUUACCAACAAUAACUUUUACCCUCCACAC____ .......................((((....))))...........((((((...((((.....))))..))))))................................ (-10.27 = -10.38 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:46 2011