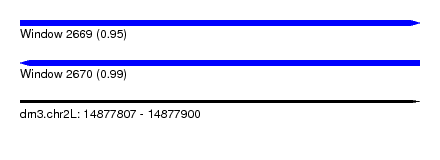
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,877,807 – 14,877,900 |
| Length | 93 |
| Max. P | 0.987712 |

| Location | 14,877,807 – 14,877,900 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.79 |
| Shannon entropy | 0.55712 |
| G+C content | 0.45019 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -10.90 |
| Energy contribution | -11.41 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
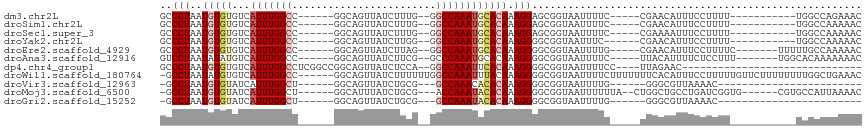
>dm3.chr2L 14877807 93 + 23011544 GCCCUAAUGUGUGUCAUUUGGCC------GGCAGUUAUCUUUG--GGCCAAAUGCACAAAGGAGCGGUAAUUUUC-----CGAACAUUUCCUUUU-----------UGGCCAGAAAC (((.......(((.(((((((((------.(.((....)).).--))))))))))))((((((((((.......)-----)).....))))))).-----------.)))....... ( -30.60, z-score = -2.32, R) >droSim1.chr2L 14627365 93 + 22036055 GCCCUAAUGUGUGUCAUUUGGCC------GGCAGUUAUCUUUG--GGCCAAAUGCACAAAGGAGCGGUAAUUUUC-----CGAACAUUUCCUUUU-----------UGGCCAAAAAC (((.......(((.(((((((((------.(.((....)).).--))))))))))))((((((((((.......)-----)).....))))))).-----------.)))....... ( -30.60, z-score = -2.46, R) >droSec1.super_3 1150230 93 + 7220098 GCCCUAAUGUGUGUCAUUUGGCC------GGCAGUUAUCUUUG--GGCCAAAUGCACAAAGGAGCGGUAAUUUUC-----CGAAAAUUUCCUUUU-----------UGGCCAAAAAC (((((....((((.(((((((((------.(.((....)).).--))))))))))))).))).))((.......(-----((((((.....))))-----------)))))...... ( -30.91, z-score = -2.61, R) >droYak2.chr2L 14983074 92 + 22324452 GCCCUAAUGUGUGUCAUUUGGCC------GGCAGUUAUCUUGG--GGCCAAAUGCACAAAGGGGCGGUAAUUUC------CGAACAUUUCCUUUU-----------UGGCCAAAAAC (((((....((((.(((((((((------...((....))...--)))))))))))))..)))))(((......------.((.....)).....-----------..)))...... ( -31.34, z-score = -1.81, R) >droEre2.scaffold_4929 6623634 97 + 26641161 GCCCUAAUGUGUGUCAUUUGGCC------GGCAGUUAUCUUAG--GGCCAAAUGCACAAAGGGGCGGUAAUUUUG-----CGAACAUUUCCUUUUC-------UUUUUGCCAAAAAC (((((....((((.(((((((((------...((....))...--)))))))))))))..)))))(((((.....-----................-------...)))))...... ( -32.26, z-score = -2.60, R) >droAna3.scaffold_12916 15207790 97 - 16180835 GUCCUAAUAUAUGUCAUUUGGCC------GGCAGUUAUCUUCG--GCCCAAAUGCACAAAGGGGCGGUAAUUUUC-----UUACAUUUUCUCCUUU-------UGGCACAAAAAAAC ((.((((....((((((((((((------((.((....)))))--).))))))).)))((((((..((((.....-----)))).....)))))))-------))).))........ ( -25.00, z-score = -2.40, R) >dp4.chr4_group1 2472377 81 - 5278887 GCCCUAAUGUGUGUCAUUUGGCCUCGGCCGGCAGUUAUCUCCA--GGCCAAAUUCACAAAGGGGCGGUAAUUUUCC----UUAGAAC------------------------------ (((((....((((..(((((((((.((..((......)).)))--)))))))).))))..)))))((.......))----.......------------------------------ ( -27.50, z-score = -2.26, R) >droWil1.scaffold_180764 3358213 110 + 3949147 -GCCUAAUAUGUGUCAUUUGGCC------GGCAGUUAUCUUUUUUGGCCAAAUUUACAAAGGGGCGGUAAUUUUCUUUUUUUCACAUUUCCUUUUUGUUCUUUUUUUUGGCUGAAAC -(((...........((((((((------((.((......)).))))))))))..(((((((((..((.................))..)))))))))..........)))...... ( -22.83, z-score = -1.17, R) >droVir3.scaffold_12963 15437075 77 - 20206255 -GCCUAAUGUGUAUCAUUUGGCU------GGCAGUUAUCUGCG---GCCAAACACACAAAGGGGCGGUAAUUUUG------GGGCGUUAAAAC------------------------ -((((..(((((....(((((((------(.(((....)))))---)))))).)))))...))))..........------............------------------------ ( -25.60, z-score = -2.52, R) >droMoj3.scaffold_6500 23659279 99 - 32352404 -GCCUAAUGUGUAUCAUUUGGCU------GGCAUUUAUCUGCG---ACCAAAUACACAAAGGGGCGGUAAUUUUUUA--CUGGCUGCCUGAUCGGUG------CGUGCCAUUAAAAC -((((..(((((((...((((.(------.(((......))).---))))))))))))...)))).......(((((--.(((((((((....)).)------)).)))).))))). ( -32.40, z-score = -2.36, R) >droGri2.scaffold_15252 660764 77 + 17193109 -GCCUAAUGUGUAUCAUUUGGCU------GGCAGUUAUCUGCG---GCCAAAUACACAAAGGGGCGGUAAUUUUG------GGGCGUUAAAAC------------------------ -((((..(((((((...((((((------(.(((....)))))---))))))))))))...))))..........------............------------------------ ( -27.60, z-score = -3.43, R) >consensus GCCCUAAUGUGUGUCAUUUGGCC______GGCAGUUAUCUUCG__GGCCAAAUGCACAAAGGGGCGGUAAUUUUC_____CGAACAUUUCCUUUU___________UGGCCAAAAAC ..(((..(((((...(((((((........................)))))))))))).)))....................................................... (-10.90 = -11.41 + 0.51)

| Location | 14,877,807 – 14,877,900 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.79 |
| Shannon entropy | 0.55712 |
| G+C content | 0.45019 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.21 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
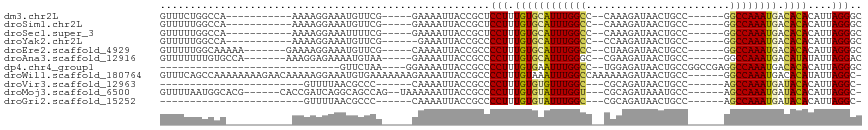
>dm3.chr2L 14877807 93 - 23011544 GUUUCUGGCCA-----------AAAAGGAAAUGUUCG-----GAAAAUUACCGCUCCUUUGUGCAUUUGGCC--CAAAGAUAACUGCC------GGCCAAAUGACACACAUUAGGGC .((((((..((-----------.........))..))-----))))......((.(((.(((((((((((((--...((....))...------))))))))).))))....))))) ( -29.40, z-score = -2.35, R) >droSim1.chr2L 14627365 93 - 22036055 GUUUUUGGCCA-----------AAAAGGAAAUGUUCG-----GAAAAUUACCGCUCCUUUGUGCAUUUGGCC--CAAAGAUAACUGCC------GGCCAAAUGACACACAUUAGGGC .......(((.-----------.((((((......((-----(.......))).))))))((((((((((((--...((....))...------))))))))).))).......))) ( -28.10, z-score = -1.86, R) >droSec1.super_3 1150230 93 - 7220098 GUUUUUGGCCA-----------AAAAGGAAAUUUUCG-----GAAAAUUACCGCUCCUUUGUGCAUUUGGCC--CAAAGAUAACUGCC------GGCCAAAUGACACACAUUAGGGC .......(((.-----------.((((((......((-----(.......))).))))))((((((((((((--...((....))...------))))))))).))).......))) ( -28.10, z-score = -2.13, R) >droYak2.chr2L 14983074 92 - 22324452 GUUUUUGGCCA-----------AAAAGGAAAUGUUCG------GAAAUUACCGCCCCUUUGUGCAUUUGGCC--CCAAGAUAACUGCC------GGCCAAAUGACACACAUUAGGGC (((((((..((-----------.........))..))------)))))....((((...(((((((((((((--...((....))...------))))))))).)))).....)))) ( -28.90, z-score = -1.78, R) >droEre2.scaffold_4929 6623634 97 - 26641161 GUUUUUGGCAAAAA-------GAAAAGGAAAUGUUCG-----CAAAAUUACCGCCCCUUUGUGCAUUUGGCC--CUAAGAUAACUGCC------GGCCAAAUGACACACAUUAGGGC .(((((......))-------)))..((.(((.....-----....))).))((((...(((((((((((((--...((....))...------))))))))).)))).....)))) ( -26.70, z-score = -1.41, R) >droAna3.scaffold_12916 15207790 97 + 16180835 GUUUUUUUGUGCCA-------AAAGGAGAAAAUGUAA-----GAAAAUUACCGCCCCUUUGUGCAUUUGGGC--CGAAGAUAACUGCC------GGCCAAAUGACAUAUAUUAGGAC ((((((((.(....-------..).))))))))((((-----.....))))....(((.(((((((((((.(--((.((....))..)------))))))))).))))....))).. ( -24.10, z-score = -1.69, R) >dp4.chr4_group1 2472377 81 + 5278887 ------------------------------GUUCUAA----GGAAAAUUACCGCCCCUUUGUGAAUUUGGCC--UGGAGAUAACUGCCGGCCGAGGCCAAAUGACACACAUUAGGGC ------------------------------.......----((.......))((((...((((.((((((((--(((.(........)..)).)))))))))....))))...)))) ( -26.40, z-score = -2.20, R) >droWil1.scaffold_180764 3358213 110 - 3949147 GUUUCAGCCAAAAAAAAGAACAAAAAGGAAAUGUGAAAAAAAGAAAAUUACCGCCCCUUUGUAAAUUUGGCCAAAAAAGAUAACUGCC------GGCCAAAUGACACAUAUUAGGC- ......(((..............(((((....((((...........))))....)))))((..((((((((.....((....))...------))))))))...))......)))- ( -18.80, z-score = -1.05, R) >droVir3.scaffold_12963 15437075 77 + 20206255 ------------------------GUUUUAACGCCC------CAAAAUUACCGCCCCUUUGUGUGUUUGGC---CGCAGAUAACUGCC------AGCCAAAUGAUACACAUUAGGC- ------------------------............------..........(((....((((((((((((---.((((....)))).------.))))...))))))))...)))- ( -22.10, z-score = -3.13, R) >droMoj3.scaffold_6500 23659279 99 + 32352404 GUUUUAAUGGCACG------CACCGAUCAGGCAGCCAG--UAAAAAAUUACCGCCCCUUUGUGUAUUUGGU---CGCAGAUAAAUGCC------AGCCAAAUGAUACACAUUAGGC- .(((((.((((..(------(.(......))).)))).--))))).......(((....((((((((((((---.(((......))).------.))))...))))))))...)))- ( -26.40, z-score = -1.55, R) >droGri2.scaffold_15252 660764 77 - 17193109 ------------------------GUUUUAACGCCC------CAAAAUUACCGCCCCUUUGUGUAUUUGGC---CGCAGAUAACUGCC------AGCCAAAUGAUACACAUUAGGC- ------------------------............------..........(((....((((((((((((---.((((....)))).------.))))...))))))))...)))- ( -22.10, z-score = -3.68, R) >consensus GUUUUUGGCCA___________AAAAGGAAAUGUUCG_____GAAAAUUACCGCCCCUUUGUGCAUUUGGCC__CGAAGAUAACUGCC______GGCCAAAUGACACACAUUAGGGC .......................................................(((.((((((((((((........................)))))))).))))....))).. (-13.40 = -13.21 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:43 2011