| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,874,764 – 14,874,889 |
| Length | 125 |
| Max. P | 0.522657 |

| Location | 14,874,764 – 14,874,889 |
|---|---|
| Length | 125 |
| Sequences | 8 |
| Columns | 133 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.52164 |
| G+C content | 0.59692 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -22.30 |
| Energy contribution | -23.35 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
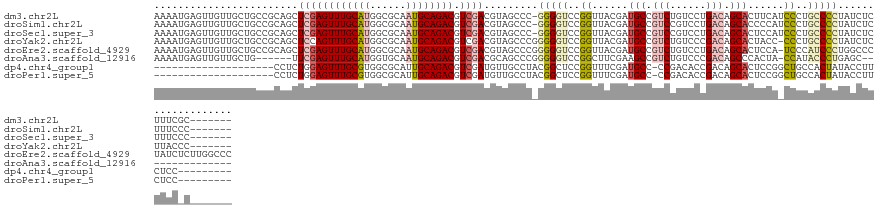
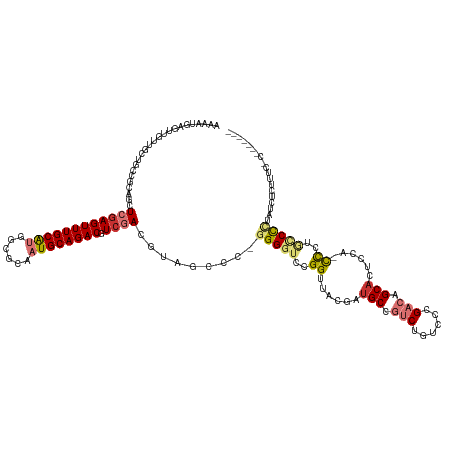
>dm3.chr2L 14874764 125 - 23011544 AAAAUGAGUUGUUGCUGCCGCAGCUCGAGUUUGCAUGGCGCAAUGCAGACGUCGACGUAGCCC-GGGGUCCGGUUACGAUGCCGUCUGUCCUGACAGCACUUCAUCCCUGCCCCUAUCUCUUUCGC------- .....(((..(((((....)))))((((((((((((......)))))))).))))........-(((((..((......(((.(((......))).)))......))..)))))...)))......------- ( -40.30, z-score = -0.54, R) >droSim1.chr2L 14624299 125 - 22036055 AAAAUGAGUUGUUGCUGCCGCAGCUCGAGUUUGCAUGGCGCAAUGCAGACGUCGACGUAGCCC-GGGGUCCGGUUACGAUGCCGUCCGUCCUGACAGCACCCCAUCCCUGCCCCUAUCUCUUUCCC------- .....(((..(((((....)))))((((((((((((......)))))))).)))).((((...-(((((.(.((((.((((.....)))).)))).).)))))....))))......)))......------- ( -39.70, z-score = -0.99, R) >droSec1.super_3 1147171 125 - 7220098 AAAAUGAGUUGUUGCUGCCGCAGCUCGAGUUUGCAUGGCGCAAUGCAGACGUCGACGUAGCCC-GGGGUCCGGUUACGAUGCCGUCCGUCCUGACAGCACUCCAUCCCUGCCCCUAUCUCUUUCCC------- .....(((..(((((....)))))((((((((((((......)))))))).))))........-(((((..((....(((((.(((......))).))).))...))..)))))...)))......------- ( -39.50, z-score = -0.80, R) >droYak2.chr2L 14980026 125 - 22324452 AAAAUGAGUUGUUGCUGCCGCAGCUCCAGUUUGCAUGGCGCAAUGCAGACGUCGACGUAGCCCGGGGGUCCGGUUACGAUGCCGUCUGUCCCGACAGCACUACC-CCCUGCCCCUAUCUCUUACCC------- .....(((((((.......)))))))..((((((((......))))))))...((.((((.(.((((((..(((......)))((((((....)))).)).)))-))).)...)))).))......------- ( -39.90, z-score = -0.65, R) >droEre2.scaffold_4929 6620574 132 - 26641161 AAAAUGAGUUGUUGCUGCCGCAGCUCGAGUUUGCAUGGCGCAAUGCAGACGUCGACGUAGCCCGGGGGUCCGGUUACGAUGCCGUCUGUCCUGACAGCACUCCA-UCCCAUCCCUGGCCCUAUCUCUUGGCCC ....((((((((.......)))))))).((((((((......))))))))(((((.((((.((((((((..((....(((((.(((......))).))).))..-.)).))))))))..))))...))))).. ( -45.40, z-score = -0.50, R) >droAna3.scaffold_12916 15204540 111 + 16180835 AAAAUGAGUUGUUGCUG------UUCGAGUUUGCAUGGUGCAAUGCAGACGUCGACGCAGCCCGGGGGUCCGGCUUCGAAGCCGUCUGUCCCGACAGCCCACUA-CCAUACCCUGAGC--------------- ....((.((((((((((------(((((((((((((......)))))))).)))).)))))..((((...(((((....)))))....)))))))))).))...-.............--------------- ( -41.30, z-score = -1.67, R) >dp4.chr4_group1 2468169 103 + 5278887 --------------------CCUCUGGAGUUUGCGUGGCGCAUUGCAGACGUCGAUGUUGCCUACGGCUCCGGUUUCGAUGCC-CCGACACCGACAGCACUCCGGCUGCCACUAUACCUUCUCC--------- --------------------...(((((((...(((((((((((((....).))))).)))).)))(((.((((.(((.....-.))).))))..))))))))))...................--------- ( -33.20, z-score = -0.97, R) >droPer1.super_5 2398459 103 - 6813705 --------------------CCUCUGGAGUUUGCGUGGCGCAUUGCAGACGUCGAUGUUGCCUACGGCUCCGGUUUCGAUGCC-CCGACACCGACAGCACUCCGGCUGCCACUAUACCUUCUCC--------- --------------------...(((((((...(((((((((((((....).))))).)))).)))(((.((((.(((.....-.))).))))..))))))))))...................--------- ( -33.20, z-score = -0.97, R) >consensus AAAAUGAGUUGUUGCUGCCGCAGCUCGAGUUUGCAUGGCGCAAUGCAGACGUCGACGUAGCCC_GGGGUCCGGUUACGAUGCCGUCUGUCCCGACAGCACUCCA_CCCUGCCCCUAUCUCUUUC_C_______ ........................((((((((((((......)))))))).)))).........(((((..((......(((.(((......))).)))......))..)))))................... (-22.30 = -23.35 + 1.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:42 2011