| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,872,919 – 14,873,024 |
| Length | 105 |
| Max. P | 0.933287 |

| Location | 14,872,919 – 14,873,024 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 55.53 |
| Shannon entropy | 0.92497 |
| G+C content | 0.32897 |
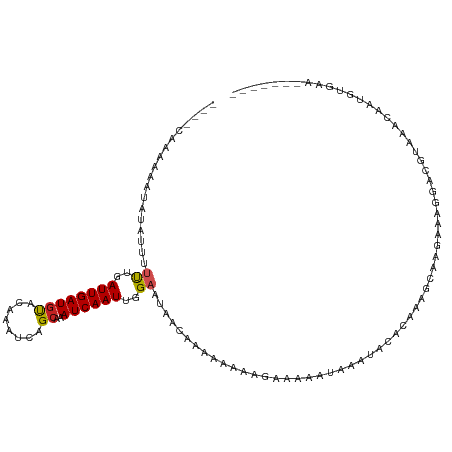
| Mean single sequence MFE | -13.19 |
| Consensus MFE | -6.59 |
| Energy contribution | -6.54 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.45 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
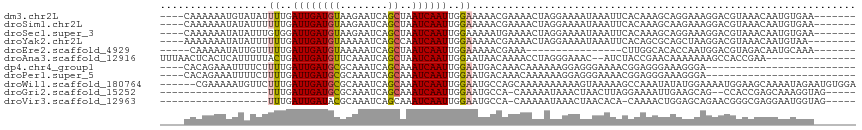
>dm3.chr2L 14872919 105 - 23011544 ----CAAAAAAUGUAUAUUUUGAUUGAUGUAAGAAUCAGCUAAUCAAUUGGAAAAACGAAAACUAGGAAAAUAAAUUCACAAAGCAGGAAAGGACGUAAACAAUGUGAA------- ----.............(((..((((((...((......)).))))))..)))......................((((((......................))))))------- ( -10.75, z-score = 0.20, R) >droSim1.chr2L 14622076 105 - 22036055 ----CAAAAAAUAUAUUUUUUGAUUGAUGUAAGAAUCAGCUAAUCAAUUGGAAAAACGAAAACUAGGAAAAUAAAUUCACAAAGCAAGAAAGGACGUAAACAAUGUGAA------- ----...........(((((..((((((...((......)).))))))..)))))....................((((((......................))))))------- ( -12.55, z-score = -0.69, R) >droSec1.super_3 1145014 105 - 7220098 ----CAAAAAAUAUAUUUGUGGAUUGAUGUAAGAAUCAGCUAAUCAAUUGGAAAAAUGAAAACUAGGAAAAUAAAUUCACAAAGCAGGAAAGGACGUAAACAAUGUGAA------- ----...........(((((((((((((......)))......(((.((....)).)))..............))))))))))..........((((.....))))...------- ( -12.80, z-score = -0.67, R) >droYak2.chr2L 14977684 104 - 22324452 ----AAAAAAAUAUAUUUUUUGAUUGAUGUAAAAAUCAGCCAAUCAAUUGGAAAAACGAAAACUAGGAAAAUAAAUUCACAGCGCAGCUAAGGACGUAAACAAUGUAA-------- ----.......(((((((((..((((((..............))))))..)))))(((........(((......)))..(((...))).....)))......)))).-------- ( -12.04, z-score = -0.01, R) >droEre2.scaffold_4929 6618323 88 - 26641161 -----CAAAAAUAUUGUUUUUGAUUGAUGUAAAAAUCAGCUAAUCAAUUGGAAAAACGAAA----------------CUUGGCACACCAAUGGACGUAGACAAUGCAAA------- -----......(((((((((..((((((..............))))))..))...(((...----------------(((((....)))).)..))).)))))))....------- ( -15.04, z-score = -1.09, R) >droAna3.scaffold_12916 15202839 99 + 16180835 UUUAACUCACUCAUUUUUACUGAUUGAUGUUCAAAUCAGCUAAUCAAUUGGAAUAACAAAACCUAGGGAAAC--AUCUACCGAACAAAAAAAGCCACCGAA--------------- ......(((.(((.......))).)))(((((...............(((......))).......(....)--.......)))))...............--------------- ( -9.60, z-score = 0.43, R) >dp4.chr4_group1 2464801 87 + 5278887 ----CACAGAAAUUUUCUUUUGAUUGAUGCGCAAAUCAGCAAAUCAAUUGGAAUGACAAACAAAAAAGGAGGGAAAACGGAGGGAAAGGGA------------------------- ----........((((((((((.(((((..((......))..)))))(((......)))..................))))))))))....------------------------- ( -13.80, z-score = -1.12, R) >droPer1.super_5 2395167 87 - 6813705 ----CACAGAAAUUUUCUUUUGAUUGAUGCGCAAAUCAGCAAAUCAAUUGGAAUGACAAACAAAAAAGGAGGGAAAACGGAGGGAAAGGGA------------------------- ----........((((((((((.(((((..((......))..)))))(((......)))..................))))))))))....------------------------- ( -13.80, z-score = -1.12, R) >droWil1.scaffold_180764 3350129 110 - 3949147 ------CGAAAAAUGUUCUUUGAUUGAUGCGCAAAUCAGCAAAUCAAUUGGAAUGCCAGCAAAAAAAAAAGUAAAAAGCCAAAUAUAUGGAAAAUGGAAGCAAAAUAGAAUGUGGA ------......((((((((..((((((..((......))..))))))..))...(((((..........))......(((......)))....)))..........))))))... ( -15.80, z-score = 0.31, R) >droGri2.scaffold_15252 653480 90 - 17193109 ------------------UUUGAUUGAUGCGCAAAUCAGCAAAUCAAUUGGAAUGCCA-CAAAAAUAAACUAACUUAGGAAAAUUGAAGCAG--CCACCGAGCAAAGGUAG----- ------------------((..((((((..((......))..))))))..)).((((.-..........(((...))).............(--(......))...)))).----- ( -14.90, z-score = -0.31, R) >droVir3.scaffold_12963 15429997 91 + 20206255 ------------------UUUGAUUGAUACGCAAAUCAGCAAAUCAAUUGGAAUGCCA-CAAAAAUAAACUAACACA-CAAAACUGGAGCAGAACGGGCGAGGAAUGGUAG----- ------------------((..((((((..((......))..))))))..)).(((((-..................-((....))..((.......))......))))).----- ( -14.00, z-score = -0.84, R) >consensus ____CAAAAAAUAUAUUUUUUGAUUGAUGUACAAAUCAGCAAAUCAAUUGGAAUAACAAAAAAAAGAAAAAUAAAUACACAAAGCAAGAAAGGACGUAAACAAUGUGAA_______ ..................((..((((((((........))..))))))..))................................................................ ( -6.59 = -6.54 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:41 2011