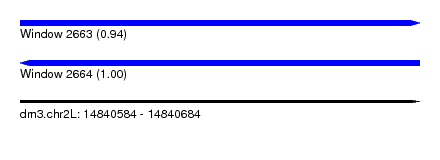
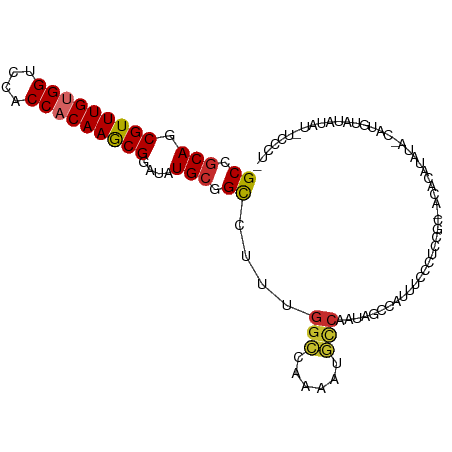
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,840,584 – 14,840,684 |
| Length | 100 |
| Max. P | 0.999461 |

| Location | 14,840,584 – 14,840,684 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 71.08 |
| Shannon entropy | 0.59427 |
| G+C content | 0.52665 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -17.33 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14840584 100 + 23011544 GAGGGA-AUGUAUAUAUG-UAUACGUGU-GCGGAGGGAAAUGGCUAUUGGCAUUUUGGCCAAAGGCCGCACAUCCGCUUGUGGUGGACCACAAACGCUGCGGC ......-..(((((....-)))))((((-(((((.(....(((((.(((((......))))).)))))..).)))))((((((....))))))))))...... ( -36.60, z-score = -1.97, R) >droSim1.chr2L 14589361 101 + 22036055 GAGGGA-AUGUAUAAAUGGUAUACGUGU-GCGGAGGGAAAUGGCUAUUGGCAUUUUGGCCAAAGGCCGCACAUCCGCUUGUGGUGGACCACAAACGCUGCGGC ......-..(((((.....)))))((((-(((((.(....(((((.(((((......))))).)))))..).)))))((((((....))))))))))...... ( -36.30, z-score = -1.83, R) >droSec1.super_3 1112956 101 + 7220098 GAGGGA-AUGUAUAAAUGGUAUACGUGU-GCGGAGGGAAAUGGCUAUUGGCAUUUUGGCCAAAGGCCGCACAUCCGCUUGUGGUGGACCACAAACGCUGCGGC ......-..(((((.....)))))((((-(((((.(....(((((.(((((......))))).)))))..).)))))((((((....))))))))))...... ( -36.30, z-score = -1.83, R) >droYak2.chr2L 14945259 97 + 22324452 AAGGGA----UAUAAAUG-UGUAUGUGU-GCGGAGUGAAAUGGCUAUUGGCAUUUUGGCCAAAGGCCGCAUAUCCGCUUGUGGUGGACCACAAACGCUGCGGC ......----........-((((.((((-((((((((....((((.(((((......))))).)))).))).)))))((((((....)))))))))))))).. ( -36.60, z-score = -2.40, R) >droEre2.scaffold_4929 6585699 101 + 26641161 AAGGGAUAUAUGUAAAUG-UAUAUGUGU-GCGGAGUGAAAUGGCUAUUGGCAUUUUGGCCAAAGGCCGCAUAUCCGCUUGUGGUAGACCGCAAACGCUGCGGC .((.((((((((((....-)))))))))-((((((((....((((.(((((......))))).)))).))).)))))((((((....)))))).).))..... ( -36.30, z-score = -2.06, R) >droAna3.scaffold_12916 15171135 97 - 16180835 --CGUCCAUACAUUCAUA-CAUAUUUGU-GUGCGGGCAGGCCGUU--UGGAAUUUCGUACAGGGGCUGCAUAUCCGUUUGUGGCGGACCACAAACGCUGCGGC --..((((.((...((((-((....)))-)))..((....)))).--)))).............((((((....(((((((((....))))))))).)))))) ( -33.00, z-score = -1.08, R) >dp4.chr4_group1 2890940 102 + 5278887 -AGAGGCAUAAGCCCAAAGAAGAUUCGUCGAGUGGGUUUUUGGGCAUUGACGUUUUGGCCUGGUGCUGCAUAUCCGCUUGUGGUGGACCACAAACGCUGCGGC -..((((....(((((((((..((((...))))...))))))))).....(.....)))))...((((((....((.((((((....)))))).)).)))))) ( -34.20, z-score = -0.45, R) >droPer1.super_5 4488839 102 - 6813705 -AGAGGCAUAAGCCCAAAGAAGAUUCGUCGAGUGGGUUUUUGGGCAUUGACGUUUUGGCCUGGUGCUGCAUAUCCGCUUGUGGUGGACCACAAACGCUGCGGC -..((((....(((((((((..((((...))))...))))))))).....(.....)))))...((((((....((.((((((....)))))).)).)))))) ( -34.20, z-score = -0.45, R) >droMoj3.scaffold_6500 23599120 84 - 32352404 ----------CACUCACA-UACGCACUC-AAAUAGGAAUGCGGGU----GAAUAUUCGC---GGACUGCAUAUCCGCUUGUGGUGCACCGCAAACGCUGUGGC ----------(((((((.-..((((.((-......)).)))).))----))......((---(((.......)))))((((((....)))))).....))).. ( -27.10, z-score = -1.39, R) >consensus _AGGGA_AUAUAUAAAUG_UAUAUGUGU_GCGGAGGGAAAUGGCUAUUGGCAUUUUGGCCAAAGGCCGCAUAUCCGCUUGUGGUGGACCACAAACGCUGCGGC ................................................................((((((....((.((((((....)))))).)).)))))) (-17.33 = -16.92 + -0.41)

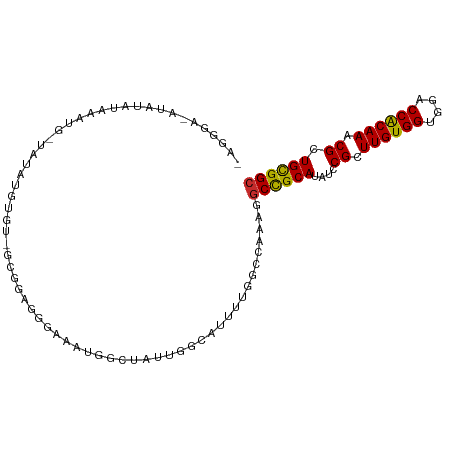
| Location | 14,840,584 – 14,840,684 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 71.08 |
| Shannon entropy | 0.59427 |
| G+C content | 0.52665 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14840584 100 - 23011544 GCCGCAGCGUUUGUGGUCCACCACAAGCGGAUGUGCGGCCUUUGGCCAAAAUGCCAAUAGCCAUUUCCCUCCGC-ACACGUAUA-CAUAUAUACAU-UCCCUC ((((((.(((((((((....)))))))))..)))).(((..(((((......)))))..)))..........))-....(((((-....)))))..-...... ( -34.50, z-score = -3.99, R) >droSim1.chr2L 14589361 101 - 22036055 GCCGCAGCGUUUGUGGUCCACCACAAGCGGAUGUGCGGCCUUUGGCCAAAAUGCCAAUAGCCAUUUCCCUCCGC-ACACGUAUACCAUUUAUACAU-UCCCUC ((((((.(((((((((....)))))))))..)))).(((..(((((......)))))..)))..........))-....(((((.....)))))..-...... ( -34.60, z-score = -4.12, R) >droSec1.super_3 1112956 101 - 7220098 GCCGCAGCGUUUGUGGUCCACCACAAGCGGAUGUGCGGCCUUUGGCCAAAAUGCCAAUAGCCAUUUCCCUCCGC-ACACGUAUACCAUUUAUACAU-UCCCUC ((((((.(((((((((....)))))))))..)))).(((..(((((......)))))..)))..........))-....(((((.....)))))..-...... ( -34.60, z-score = -4.12, R) >droYak2.chr2L 14945259 97 - 22324452 GCCGCAGCGUUUGUGGUCCACCACAAGCGGAUAUGCGGCCUUUGGCCAAAAUGCCAAUAGCCAUUUCACUCCGC-ACACAUACA-CAUUUAUA----UCCCUU ((.(((.(((((((((....)))))))))....)))(((..(((((......)))))..)))..........))-.........-........----...... ( -31.60, z-score = -3.85, R) >droEre2.scaffold_4929 6585699 101 - 26641161 GCCGCAGCGUUUGCGGUCUACCACAAGCGGAUAUGCGGCCUUUGGCCAAAAUGCCAAUAGCCAUUUCACUCCGC-ACACAUAUA-CAUUUACAUAUAUCCCUU (((((((...))))))).........(((((.....(((..(((((......)))))..))).......)))))-.........-.................. ( -27.70, z-score = -2.38, R) >droAna3.scaffold_12916 15171135 97 + 16180835 GCCGCAGCGUUUGUGGUCCGCCACAAACGGAUAUGCAGCCCCUGUACGAAAUUCCA--AACGGCCUGCCCGCAC-ACAAAUAUG-UAUGAAUGUAUGGACG-- ((.(((.(((((((((....)))))))))....))).))..(((((((........--..(((.....)))((.-(((....))-).))..)))))))...-- ( -27.00, z-score = -0.49, R) >dp4.chr4_group1 2890940 102 - 5278887 GCCGCAGCGUUUGUGGUCCACCACAAGCGGAUAUGCAGCACCAGGCCAAAACGUCAAUGCCCAAAAACCCACUCGACGAAUCUUCUUUGGGCUUAUGCCUCU- ((.(((.(((((((((....)))))))))....))).))...((((............(((((((....................)))))))....))))..- ( -30.24, z-score = -1.61, R) >droPer1.super_5 4488839 102 + 6813705 GCCGCAGCGUUUGUGGUCCACCACAAGCGGAUAUGCAGCACCAGGCCAAAACGUCAAUGCCCAAAAACCCACUCGACGAAUCUUCUUUGGGCUUAUGCCUCU- ((.(((.(((((((((....)))))))))....))).))...((((............(((((((....................)))))))....))))..- ( -30.24, z-score = -1.61, R) >droMoj3.scaffold_6500 23599120 84 + 32352404 GCCACAGCGUUUGCGGUGCACCACAAGCGGAUAUGCAGUCC---GCGAAUAUUC----ACCCGCAUUCCUAUUU-GAGUGCGUA-UGUGAGUG---------- ......(((((((.((....)).)))))((((.....))))---))...(((((----((.(((((((......-)))))))..-.)))))))---------- ( -29.30, z-score = -2.17, R) >consensus GCCGCAGCGUUUGUGGUCCACCACAAGCGGAUAUGCGGCCUUUGGCCAAAAUGCCAAUAGCCAUUUCCCUCCGC_ACACAUAUA_CAUGUAUAUAU_UCCCU_ ((.(((.(((((((((....)))))))))....))).))....(((......)))................................................ (-18.33 = -18.40 + 0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:38 2011