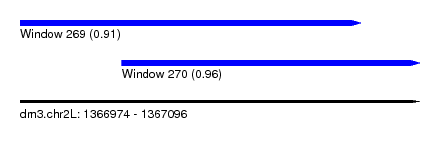
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,366,974 – 1,367,096 |
| Length | 122 |
| Max. P | 0.963650 |

| Location | 1,366,974 – 1,367,078 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.92 |
| Shannon entropy | 0.35785 |
| G+C content | 0.52450 |
| Mean single sequence MFE | -17.33 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.73 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
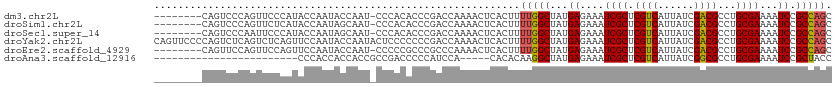
>dm3.chr2L 1366974 104 + 23011544 --------CAGUCCCAGUUCCCAUACCAAUACCAAU-CCCACACCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC --------.........(((.((((((((.......-........................)))).)))).))).((((.((((......))))...))))............ ( -15.98, z-score = -1.33, R) >droSim1.chr2L 1340307 104 + 22036055 --------CAGUCCCAGUUCUCAUACCAAUAGCAAU-CCCACACCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC --------.........((((((((((((.((....-.....................)).)))).)))))))).((((.((((......))))...))))............ ( -21.61, z-score = -2.67, R) >droSec1.super_14 1314966 104 + 2068291 --------CAGUCCCAAUUCCCAUACCAAUAGCAAU-CCCACACCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC --------.........(((.((((((((.((....-.....................)).)))).)))).))).((((.((((......))))...))))............ ( -16.91, z-score = -1.55, R) >droYak2.chr2L 1342178 113 + 22324452 CAGUUCCCCAGUCUCAGUCUCAGUUCCAAUACCAAUACUCCCCCCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC .................(((((((......))..............(.((((((.....)))))))..)))))..((((.((((......))))...))))............ ( -17.80, z-score = -1.00, R) >droEre2.scaffold_4929 1410903 104 + 26641161 --------CAGUUCCAGUUCCAGUUCCAAUACCAAU-CCCCCGCCCGCCCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC --------............................-........................(((((...((....((((.((((......))))...))))...)).))))). ( -15.60, z-score = -0.28, R) >droAna3.scaffold_12916 4828899 84 + 16180835 ------------------------CCCACCACCACCGCCGACCCCCAUCCA-----CACACAAGGCUAUGAGAAAUCGCUCGUCAUUAUCGGCGCCUGCGAAAAUCCGCUACC ------------------------...........((((((.......((.-----.......))..(((((......))))).....))))))...(((......))).... ( -16.10, z-score = -1.52, R) >consensus ________CAGUCCCAGUUCCCAUACCAAUACCAAU_CCCACACCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC .............................................................(((((...((....((((.((((......))))...))))...)).))))). (-14.68 = -14.73 + 0.06)
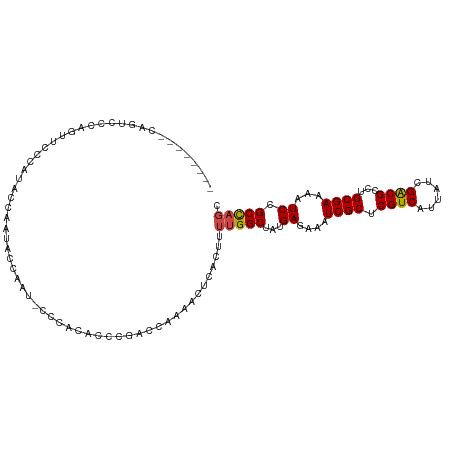
| Location | 1,367,005 – 1,367,096 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Shannon entropy | 0.44160 |
| G+C content | 0.51994 |
| Mean single sequence MFE | -18.07 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.33 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1367005 91 + 23011544 -ACACCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAG-CUCAGACACACAGCCACAU------------ -......................(((((..((....))(((((((......))))...(((......))).))-)..........)))))...------------ ( -17.20, z-score = -1.50, R) >droSim1.chr2L 1340338 91 + 22036055 -ACACCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAG-CUCAGACACACACCCACAU------------ -.....................(((((...((....((((.((((......))))...))))...)).)))))-...................------------ ( -15.60, z-score = -1.57, R) >droSec1.super_14 1314997 91 + 2068291 -ACACCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAG-CUCAGACACACACCCACAU------------ -.....................(((((...((....((((.((((......))))...))))...)).)))))-...................------------ ( -15.60, z-score = -1.57, R) >droYak2.chr2L 1342218 91 + 22324452 -CCCCCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAG-CUCAGACACACACUCACAU------------ -......(.((((((.....)))))))..((((.........(((......)))((..(((......)))..)-)...........))))...------------ ( -16.00, z-score = -1.60, R) >droEre2.scaffold_4929 1410934 89 + 26641161 -CCGCCCGCCCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAG-CUCAGACACAC--UCACAU------------ -..((..((((((((.....))))))..........((((.((((......))))...))))......))..)-)..........--......------------ ( -16.70, z-score = -1.03, R) >droAna3.scaffold_12916 4828919 93 + 16180835 ----------CCAUCCACACACAAGGCUAUGAGAAAUCGCUCGUCAUUAUCGGCGCCUGCGAAAAUCCGCUA--CCCACCCACACGCACCCAUCCACAUGUCCUU ----------............((((.((((.....((((.((((......))))...))))......((..--...........)).........)))).)))) ( -15.52, z-score = -0.86, R) >droVir3.scaffold_12963 7324998 83 + 20206255 CCACCGAGUGCGACGAGUGGCGCUGGCUAUGAGAAAUCGCUCGUCAUUAUUGACGCUUGCGAAAAUUCGCUAGAAUCGCACAC---------------------- .......((((((((((((((....)))..((....)))))))))......((..((.((((....)))).))..))))))..---------------------- ( -29.90, z-score = -2.38, R) >consensus _ACACCCGACCAAAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAG_CUCAGACACACACCCACAU____________ ......................(((((...((....((((.((((......))))...))))...)).)))))................................ (-14.59 = -14.33 + -0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:24 2011