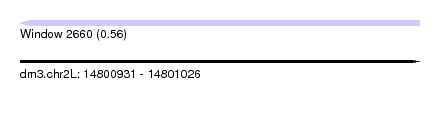
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,800,931 – 14,801,026 |
| Length | 95 |
| Max. P | 0.563670 |

| Location | 14,800,931 – 14,801,026 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Shannon entropy | 0.33057 |
| G+C content | 0.49501 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -13.19 |
| Energy contribution | -13.11 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
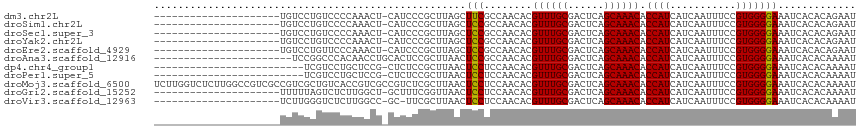
>dm3.chr2L 14800931 95 - 23011544 ---------------------UGUCCUGUCCCCAAACU-CAUCCCGCUUAGCUUCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAU ---------------------(((....((((((....-......((........))......((((((......)))))).................))))))....)))...... ( -20.60, z-score = -2.04, R) >droSim1.chr2L 14553846 95 - 22036055 ---------------------UGUCCUGUCCCCAAACU-CAUCCCGCUUAGCUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAU ---------------------(((....((((((....-......((........))......((((((......)))))).................))))))....)))...... ( -20.60, z-score = -2.00, R) >droSec1.super_3 1078043 95 - 7220098 ---------------------UGUCCUGUCCCCAAACU-CAUCCCGCUUAGCUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAU ---------------------(((....((((((....-......((........))......((((((......)))))).................))))))....)))...... ( -20.60, z-score = -2.00, R) >droYak2.chr2L 14908781 95 - 22324452 ---------------------UGUCCUGUCCCCAAACU-CAUCCCGCUUAGCUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAU ---------------------(((....((((((....-......((........))......((((((......)))))).................))))))....)))...... ( -20.60, z-score = -2.00, R) >droEre2.scaffold_4929 6548325 95 - 26641161 ---------------------UGUCCUGUUCCCAAACU-CAUCCCGCUUAGCUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAU ---------------------(((....((((((....-......((........))......((((((......)))))).................))))))....)))...... ( -17.70, z-score = -0.96, R) >droAna3.scaffold_12916 15129701 94 + 16180835 -----------------------UCCGGCCCACAACCUGCACUCCGCUUAACUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAU -----------------------.....(((((.....((.....))................((((((......))))))................)))))............... ( -18.60, z-score = -1.67, R) >dp4.chr4_group1 2333018 91 + 5278887 -------------------------UCGUCCUGCUCCG-CUCUCCGCUUAACUCCUCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAU -------------------------...(((..(...(-(.....))................((((((......))))))................)..))).............. ( -15.60, z-score = -1.57, R) >droPer1.super_5 2271303 91 - 6813705 -------------------------UCGUCCUGCUCCG-CUCUCCGCUUAACUCCUCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAU -------------------------...(((..(...(-(.....))................((((((......))))))................)..))).............. ( -15.60, z-score = -1.57, R) >droMoj3.scaffold_6500 23541507 117 + 32352404 UCUUGGUCUCUUGGCCGUCGCCGUCGCUGUCACCGUCGCCGUCUCGCUUAACUCCUCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAU ....((((....))))(.((.((.((.......)).)).)).)............(((.....((((((......)))))).((((...........)))))))............. ( -23.50, z-score = 0.15, R) >droGri2.scaffold_15252 558353 95 - 17193109 ---------------------UUUUUAGUCUCUUGGCU-GCUUUCGGUUAACUCCUCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAU ---------------------....(((((....))))-).....((......))(((.....((((((......)))))).((((...........)))))))............. ( -18.20, z-score = -1.21, R) >droVir3.scaffold_12963 15285918 94 + 20206255 ---------------------UCUUGGGUCUCUUGGCC-GC-UUCGCUUAACUCCUCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAU ---------------------..(((((((....))))-..-.............(((.....((((((......)))))).((((...........))))))).......)))... ( -19.60, z-score = -1.21, R) >consensus _____________________UGUCCUGUCCCCAAACU_CAUCCCGCUUAACUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAU ....................................................(((........((((((......)))))).((((...........)))))))............. (-13.19 = -13.11 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:35 2011