| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,789,864 – 14,789,921 |
| Length | 57 |
| Max. P | 0.962048 |

| Location | 14,789,864 – 14,789,921 |
|---|---|
| Length | 57 |
| Sequences | 10 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Shannon entropy | 0.47734 |
| G+C content | 0.52346 |
| Mean single sequence MFE | -17.57 |
| Consensus MFE | -10.84 |
| Energy contribution | -10.66 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.921317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
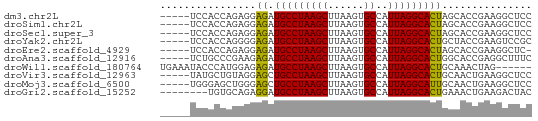
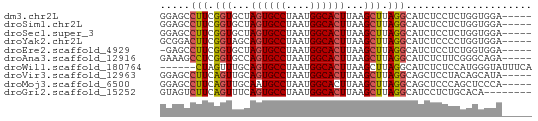
>dm3.chr2L 14789864 57 + 23011544 -----UCCACCAGAGGAGAUGCCUAAGCUUAAGUGCCAUUAGGCACUAGCACCGAAGGCUCC -----(((......)))((.((((..(((..((((((....))))))))).....)))))). ( -18.90, z-score = -2.35, R) >droSim1.chr2L 14540173 57 + 22036055 -----UCCACCAGAGGAGAUGCCUAAGCUUAAGUGCCAUUAGGCACUAGCACCGAAGGCUCC -----(((......)))((.((((..(((..((((((....))))))))).....)))))). ( -18.90, z-score = -2.35, R) >droSec1.super_3 1066883 57 + 7220098 -----UCCACCAGAGGAGAUGCCUAAGCUUAAGUGCCAUUAGGCACUAGCACCGAAGGCUCC -----(((......)))((.((((..(((..((((((....))))))))).....)))))). ( -18.90, z-score = -2.35, R) >droYak2.chr2L 14898916 57 + 22324452 -----UCCACCAGGGGAGAUGCCUAAGCUUAAGUGCCAUUAGGCACUGCUACCGAAGUCCGC -----(((......)))(((.....(((...((((((....)))))))))......)))... ( -14.20, z-score = 0.17, R) >droEre2.scaffold_4929 6537083 56 + 26641161 -----UCCACCAGAGGAGAUGCCUAAGCUUAAGUGCCAUUAGGCACUAGCACCGAAGGCUC- -----(((......)))...((((..(((..((((((....))))))))).....))))..- ( -18.40, z-score = -2.24, R) >droAna3.scaffold_12916 15120159 57 - 16180835 -----UCUGCCCGAAGAGAUGCCUAAGCUUAAGUGCCAUUAGGCACUGGCACCGAGGCUUUC -----..........((((.((((..(((..((((((....)))))))))....)))))))) ( -19.70, z-score = -1.90, R) >droWil1.scaffold_180764 3238689 56 + 3949147 UGAAAUACCCAUGGAGAGAUGCCUAAGCUUAAGUGCCAUUAGGCACUGCAAACUAG------ ...........((.((...(((((((((......))..))))))))).))......------ ( -11.90, z-score = -1.03, R) >droVir3.scaffold_12963 15259968 57 - 20206255 -----UAUGCUGUAGGAGCUGCCUAAGCUUAAGUGCCAUUAGGCACUGCAACUGAAGGCUCC -----.........((((((.....((....((((((....))))))....))...)))))) ( -20.20, z-score = -1.32, R) >droMoj3.scaffold_6500 23520538 57 - 32352404 -----UGGGAGCUGGGAGCUGCCUAAGCUUAAGUGCCAUUAGGCAUUGCAACUGAAGGCUCC -----..((((((..(((((.....))))).((((((....)))))).........)))))) ( -21.70, z-score = -1.17, R) >droGri2.scaffold_15252 541619 54 + 17193109 --------UGUGCAGAGGAUGCCUAAGCUUAAGUGCCAUUAGGCACUGAAACUGAAGACUAC --------....(((.((.(((((((((......))..)))))))))....)))........ ( -12.90, z-score = -1.08, R) >consensus _____UCCACCAGAGGAGAUGCCUAAGCUUAAGUGCCAUUAGGCACUGGCACCGAAGGCUCC ................((.(((((((((......))..)))))))))............... (-10.84 = -10.66 + -0.18)
| Location | 14,789,864 – 14,789,921 |
|---|---|
| Length | 57 |
| Sequences | 10 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Shannon entropy | 0.47734 |
| G+C content | 0.52346 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -12.49 |
| Energy contribution | -12.42 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
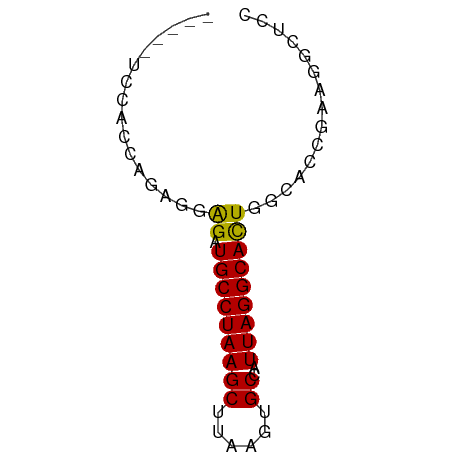
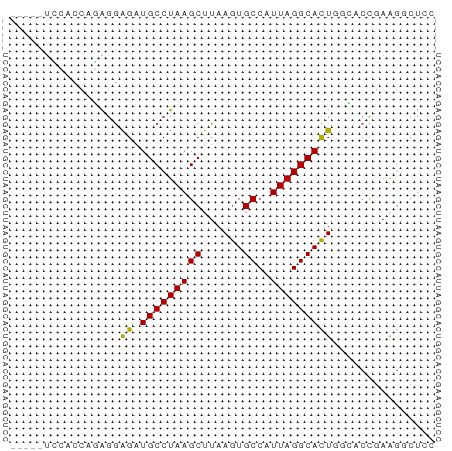
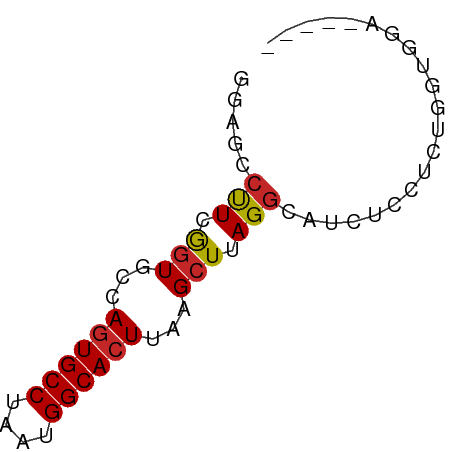
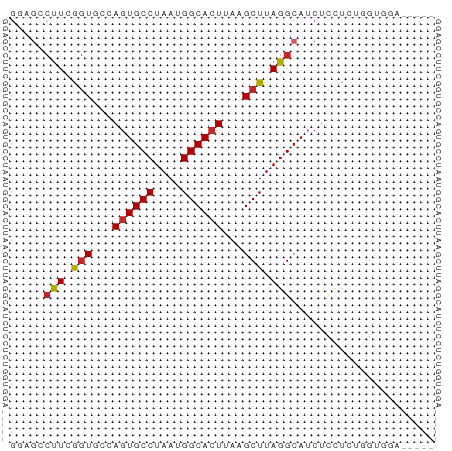
>dm3.chr2L 14789864 57 - 23011544 GGAGCCUUCGGUGCUAGUGCCUAAUGGCACUUAAGCUUAGGCAUCUCCUCUGGUGGA----- ...((((..(((...((((((....))))))...))).))))...(((......)))----- ( -21.90, z-score = -1.61, R) >droSim1.chr2L 14540173 57 - 22036055 GGAGCCUUCGGUGCUAGUGCCUAAUGGCACUUAAGCUUAGGCAUCUCCUCUGGUGGA----- ...((((..(((...((((((....))))))...))).))))...(((......)))----- ( -21.90, z-score = -1.61, R) >droSec1.super_3 1066883 57 - 7220098 GGAGCCUUCGGUGCUAGUGCCUAAUGGCACUUAAGCUUAGGCAUCUCCUCUGGUGGA----- ...((((..(((...((((((....))))))...))).))))...(((......)))----- ( -21.90, z-score = -1.61, R) >droYak2.chr2L 14898916 57 - 22324452 GCGGACUUCGGUAGCAGUGCCUAAUGGCACUUAAGCUUAGGCAUCUCCCCUGGUGGA----- ((..((....)).))((((((....))))))..........((((......))))..----- ( -17.80, z-score = -0.14, R) >droEre2.scaffold_4929 6537083 56 - 26641161 -GAGCCUUCGGUGCUAGUGCCUAAUGGCACUUAAGCUUAGGCAUCUCCUCUGGUGGA----- -..((((..(((...((((((....))))))...))).))))...(((......)))----- ( -21.90, z-score = -2.02, R) >droAna3.scaffold_12916 15120159 57 + 16180835 GAAAGCCUCGGUGCCAGUGCCUAAUGGCACUUAAGCUUAGGCAUCUCUUCGGGCAGA----- ....((((.((((((((((((....))))))........)))))).....))))...----- ( -22.00, z-score = -1.97, R) >droWil1.scaffold_180764 3238689 56 - 3949147 ------CUAGUUUGCAGUGCCUAAUGGCACUUAAGCUUAGGCAUCUCUCCAUGGGUAUUUCA ------((((((((.((((((....))))))))))).)))....(((.....)))....... ( -15.70, z-score = -1.40, R) >droVir3.scaffold_12963 15259968 57 + 20206255 GGAGCCUUCAGUUGCAGUGCCUAAUGGCACUUAAGCUUAGGCAGCUCCUACAGCAUA----- ((((((((.((((..((((((....))))))..)))).)))..))))).........----- ( -22.60, z-score = -2.25, R) >droMoj3.scaffold_6500 23520538 57 + 32352404 GGAGCCUUCAGUUGCAAUGCCUAAUGGCACUUAAGCUUAGGCAGCUCCCAGCUCCCA----- (((((....((((((..((((....))))((.......))))))))....)))))..----- ( -21.10, z-score = -1.96, R) >droGri2.scaffold_15252 541619 54 - 17193109 GUAGUCUUCAGUUUCAGUGCCUAAUGGCACUUAAGCUUAGGCAUCCUCUGCACA-------- ...((((..(((((.((((((....)))))).))))).))))............-------- ( -18.70, z-score = -3.63, R) >consensus GGAGCCUUCGGUGCCAGUGCCUAAUGGCACUUAAGCUUAGGCAUCUCCUCUGGUGGA_____ .....(((.(((...((((((....))))))...))).)))..................... (-12.49 = -12.42 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:33 2011