| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,783,057 – 14,783,109 |
| Length | 52 |
| Max. P | 0.996663 |

| Location | 14,783,057 – 14,783,109 |
|---|---|
| Length | 52 |
| Sequences | 7 |
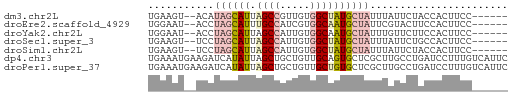
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.55854 |
| G+C content | 0.42601 |
| Mean single sequence MFE | -16.73 |
| Consensus MFE | -5.84 |
| Energy contribution | -7.19 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
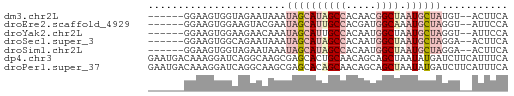
>dm3.chr2L 14783057 52 + 23011544 ------GGAAGUGGUAGAAUAAAUAGCAUAGCCACAACGGCUAAUGCUAUGU--ACUUCA ------.((((((.........(((((((((((.....)))).))))))).)--))))). ( -17.80, z-score = -3.84, R) >droEre2.scaffold_4929 6531612 52 + 26641161 ------GGAAGUGGAAGUACGAAUAGCAUUGCCACGAUGGCAAAUGCUAGGU--AUUCCA ------.....(((((.(((...((((((((((.....))).))))))).))--)))))) ( -19.00, z-score = -3.79, R) >droYak2.chr2L 14891991 52 + 22324452 ------GGAAGUGGAAGAACAAAUAGCAUUGCCACAAUGGCUAAUGCUAGGU--AUUCCA ------.....(((((..((...((((((((((.....))).))))))).))--.))))) ( -19.00, z-score = -3.92, R) >droSec1.super_3 1060094 52 + 7220098 ------GGAAGUGGCAGAAUAAAUAGCAUAGCCACAAUGGCUAAUGCUAGGA--ACUUCA ------.(((((...........((((((((((.....)))).))))))...--))))). ( -16.74, z-score = -3.25, R) >droSim1.chr2L 14533301 52 + 22036055 ------GGAAGUGGUAGAAUAAAUAGCAUAGCCACAAUGGCUAAUGCUAGGA--ACUUCA ------.(((((...........((((((((((.....)))).))))))...--))))). ( -16.74, z-score = -3.54, R) >dp4.chr3 173058 60 - 19779522 GAAUGACAAAGGAUCAGGCAAGCGAGCACUGCAACAGCAGCUAAUAUGAUCUUCAUUUCA ((((((....((((((.((......)).((((....))))......)))))))))))).. ( -14.50, z-score = -1.62, R) >droPer1.super_37 152556 60 - 760358 GAAUGACAAAGGAUCAGGCAAGCGAGCACAGCAACAGCAGCUAAUAUGAUCUUCAUUUCA ((((((....((((((....(((..((.........)).)))....)))))))))))).. ( -13.30, z-score = -1.78, R) >consensus ______GGAAGUGGCAGAAUAAAUAGCAUAGCCACAAUGGCUAAUGCUAGGU__AUUUCA .......................((((((((((.....)))).))))))........... ( -5.84 = -7.19 + 1.35)
| Location | 14,783,057 – 14,783,109 |
|---|---|
| Length | 52 |
| Sequences | 7 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.55854 |
| G+C content | 0.42601 |
| Mean single sequence MFE | -14.67 |
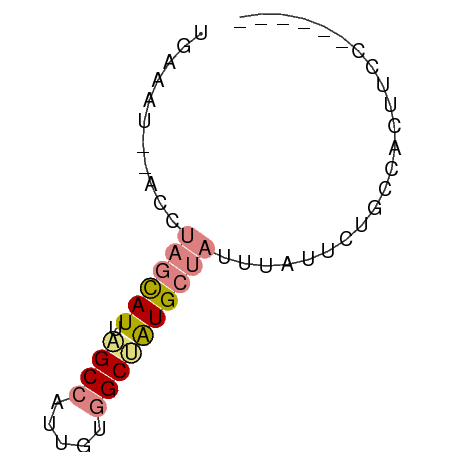
| Consensus MFE | -6.87 |
| Energy contribution | -7.73 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.996663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
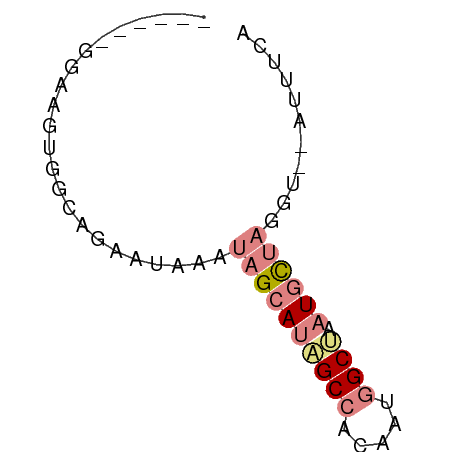
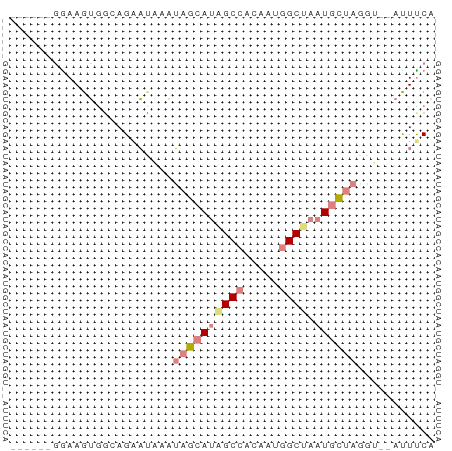
>dm3.chr2L 14783057 52 - 23011544 UGAAGU--ACAUAGCAUUAGCCGUUGUGGCUAUGCUAUUUAUUCUACCACUUCC------ ...(((--(.(((((((.((((.....))))))))))).))))...........------ ( -14.40, z-score = -2.91, R) >droEre2.scaffold_4929 6531612 52 - 26641161 UGGAAU--ACCUAGCAUUUGCCAUCGUGGCAAUGCUAUUCGUACUUCCACUUCC------ ((((((--((.(((((((.(((.....))))))))))...))).))))).....------ ( -18.20, z-score = -4.83, R) >droYak2.chr2L 14891991 52 - 22324452 UGGAAU--ACCUAGCAUUAGCCAUUGUGGCAAUGCUAUUUGUUCUUCCACUUCC------ .(((((--(..(((((((.(((.....))))))))))..)))))).........------ ( -17.50, z-score = -3.99, R) >droSec1.super_3 1060094 52 - 7220098 UGAAGU--UCCUAGCAUUAGCCAUUGUGGCUAUGCUAUUUAUUCUGCCACUUCC------ .(((..--...((((((.((((.....))))))))))....)))..........------ ( -13.80, z-score = -2.51, R) >droSim1.chr2L 14533301 52 - 22036055 UGAAGU--UCCUAGCAUUAGCCAUUGUGGCUAUGCUAUUUAUUCUACCACUUCC------ .(((..--...((((((.((((.....))))))))))....)))..........------ ( -13.80, z-score = -3.13, R) >dp4.chr3 173058 60 + 19779522 UGAAAUGAAGAUCAUAUUAGCUGCUGUUGCAGUGCUCGCUUGCCUGAUCCUUUGUCAUUC (((...((.(((((.....(((((....)))))((......)).))))).))..)))... ( -14.30, z-score = -1.19, R) >droPer1.super_37 152556 60 + 760358 UGAAAUGAAGAUCAUAUUAGCUGCUGUUGCUGUGCUCGCUUGCCUGAUCCUUUGUCAUUC (((...((.(((((....(((.((.........))..)))....))))).))..)))... ( -10.70, z-score = -0.25, R) >consensus UGAAAU__ACCUAGCAUUAGCCAUUGUGGCUAUGCUAUUUAUUCUGCCACUUCC______ ...........((((((.((((.....))))))))))....................... ( -6.87 = -7.73 + 0.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:29 2011