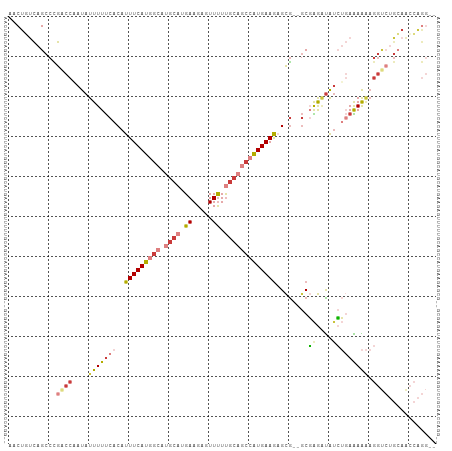
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,767,528 – 14,767,631 |
| Length | 103 |
| Max. P | 0.893220 |

| Location | 14,767,528 – 14,767,631 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.46 |
| Shannon entropy | 0.49892 |
| G+C content | 0.44238 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -13.05 |
| Energy contribution | -15.43 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14767528 103 + 23011544 AACUGUCAGCCCGACCUAUAUUUUUCACAUUUCAUGGCAUGCAUGAAGAGUUUUUGCAGCCAUGAAGAGCG--GCGAGUUAUCUGAAAAAAGGUCUGCAACCAGG-- ..(((...((..(((((...(((((((..(((((((((.((((...........)))))))))))))(((.--....)))...)))))))))))).))...))).-- ( -31.80, z-score = -2.49, R) >droSim1.chr2L 14514453 103 + 22036055 AACUGUCAGCCCGACCAAUAUUUUUCACAUUUCAUGGCAUGCAUGAAGAGUUUUUGCAGCCAUGAAGAGCG--GCGAGAUAUCUGAAAAAAGGUCUGCAACCAGG-- ..(((...((..((((....(((((((..(((((((((.((((...........)))))))))))))....--..((....))))))))).)))).))...))).-- ( -29.00, z-score = -1.77, R) >droSec1.super_3 1043591 103 + 7220098 AACUGUCAGCCCGACCAAUAUUUUUCACAUUUCAUGGCAUGCAUGAAGAGUUUUUGCAGCCAUGAAGAGCG--GCGAGAUAUCUGAAAAAAGGUCUGCAACCAGG-- ..(((...((..((((....(((((((..(((((((((.((((...........)))))))))))))....--..((....))))))))).)))).))...))).-- ( -29.00, z-score = -1.77, R) >droYak2.chr2L 14875327 103 + 22324452 AACUUUCAGCCCGACCAAUAUUUUUCACAUUUCAUGGCAUGCAUGAAGAGUUUUUGCAGCCAUGAAGAGCG--GCGAGAUAUCUGAAAAAAGGUCUGCAACCAGG-- ........((..((((....(((((((..(((((((((.((((...........)))))))))))))....--..((....))))))))).)))).)).......-- ( -26.90, z-score = -1.40, R) >droEre2.scaffold_4929 6516465 103 + 26641161 AACUGUCAGCCCGGCCAAUAUUUUUCACAUUUCAUGGCAUGCAUGAAGAGUUUUUGCAGCCAUGAAAAGCG--GCGAGAUAUCUGAAAAAAGGUCUGCAACCAGG-- ..(((...((..((((....(((((((..(((((((((.((((...........)))))))))))))....--..((....))))))))).)))).))...))).-- ( -28.70, z-score = -1.28, R) >droVir3.scaffold_12963 15235091 82 - 20206255 --CUUCUAGCCUAUCUCAUAUUUUCCACAUUUCAUAGCAUGCAUGAAGACUUCUCGCCC--AUGAAGUGUA---CUGAGUGUGGCAGAG------------------ --..(((.(((...((((...........((((((.(....)))))))(((((......--..)))))...---.))))...)))))).------------------ ( -19.00, z-score = -0.90, R) >droMoj3.scaffold_6500 23490933 99 - 32352404 --CUUCUAGCCUAUCCAAUAUUUUUCACAUUUCAUAG------UGAAGUCUUUUUGGCCCAGUGAAGUGUGUGGCUGAGCAAAGCAGAGGAGGCGAGAAGGAGAAUU --(((((.(((..(((.........((((((((((.(------(.(((....))).))...))))))))))..(((......)))...)))))).)))))....... ( -29.70, z-score = -1.80, R) >consensus AACUGUCAGCCCGACCAAUAUUUUUCACAUUUCAUGGCAUGCAUGAAGAGUUUUUGCAGCCAUGAAGAGCG__GCGAGAUAUCUGAAAAAAGGUCUGCAACCAGG__ ............((((....(((((((..(((((((((.((((.((.....)).))))))))))))).((...))........))))))).))))............ (-13.05 = -15.43 + 2.37)


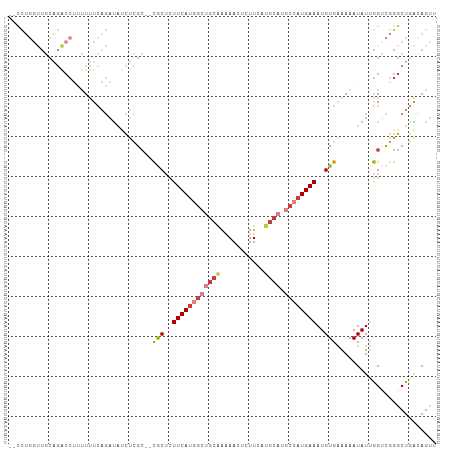
| Location | 14,767,528 – 14,767,631 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.46 |
| Shannon entropy | 0.49892 |
| G+C content | 0.44238 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -12.37 |
| Energy contribution | -13.51 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14767528 103 - 23011544 --CCUGGUUGCAGACCUUUUUUCAGAUAACUCGC--CGCUCUUCAUGGCUGCAAAAACUCUUCAUGCAUGCCAUGAAAUGUGAAAAAUAUAGGUCGGGCUGACAGUU --.(((...((.(((((((((((.((....))..--.((..((((((((((((...........)))).))))))))..))))))))...)))))..))...))).. ( -32.70, z-score = -2.84, R) >droSim1.chr2L 14514453 103 - 22036055 --CCUGGUUGCAGACCUUUUUUCAGAUAUCUCGC--CGCUCUUCAUGGCUGCAAAAACUCUUCAUGCAUGCCAUGAAAUGUGAAAAAUAUUGGUCGGGCUGACAGUU --.(((...((.((((.((((((.((....))..--.((..((((((((((((...........)))).))))))))..))))))))....))))..))...))).. ( -32.20, z-score = -2.63, R) >droSec1.super_3 1043591 103 - 7220098 --CCUGGUUGCAGACCUUUUUUCAGAUAUCUCGC--CGCUCUUCAUGGCUGCAAAAACUCUUCAUGCAUGCCAUGAAAUGUGAAAAAUAUUGGUCGGGCUGACAGUU --.(((...((.((((.((((((.((....))..--.((..((((((((((((...........)))).))))))))..))))))))....))))..))...))).. ( -32.20, z-score = -2.63, R) >droYak2.chr2L 14875327 103 - 22324452 --CCUGGUUGCAGACCUUUUUUCAGAUAUCUCGC--CGCUCUUCAUGGCUGCAAAAACUCUUCAUGCAUGCCAUGAAAUGUGAAAAAUAUUGGUCGGGCUGAAAGUU --.((....((.((((.((((((.((....))..--.((..((((((((((((...........)))).))))))))..))))))))....))))..))....)).. ( -29.10, z-score = -1.83, R) >droEre2.scaffold_4929 6516465 103 - 26641161 --CCUGGUUGCAGACCUUUUUUCAGAUAUCUCGC--CGCUUUUCAUGGCUGCAAAAACUCUUCAUGCAUGCCAUGAAAUGUGAAAAAUAUUGGCCGGGCUGACAGUU --((((((((((((..((......))..))).))--(((.(((((((((((((...........)))).))))))))).))).........)))))))......... ( -30.60, z-score = -1.89, R) >droVir3.scaffold_12963 15235091 82 + 20206255 ------------------CUCUGCCACACUCAG---UACACUUCAU--GGGCGAGAAGUCUUCAUGCAUGCUAUGAAAUGUGGAAAAUAUGAGAUAGGCUAGAAG-- ------------------.((((((...((((.---((((.(((((--(((((.((.....)).)))...))))))).)))).......))))...))).)))..-- ( -19.70, z-score = -0.55, R) >droMoj3.scaffold_6500 23490933 99 + 32352404 AAUUCUCCUUCUCGCCUCCUCUGCUUUGCUCAGCCACACACUUCACUGGGCCAAAAAGACUUCA------CUAUGAAAUGUGAAAAAUAUUGGAUAGGCUAGAAG-- .......(((((.(((((((((..((.((((((............)))))).))..))).((((------(........))))).......)))..))).)))))-- ( -25.20, z-score = -2.59, R) >consensus __CCUGGUUGCAGACCUUUUUUCAGAUAUCUCGC__CGCUCUUCAUGGCUGCAAAAACUCUUCAUGCAUGCCAUGAAAUGUGAAAAAUAUUGGUCGGGCUGACAGUU ....................................(((..((((((((((((...........)))).))))))))..)))......................... (-12.37 = -13.51 + 1.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:27 2011