| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,754,250 – 14,754,345 |
| Length | 95 |
| Max. P | 0.900210 |

| Location | 14,754,250 – 14,754,345 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.55 |
| Shannon entropy | 0.32996 |
| G+C content | 0.39942 |
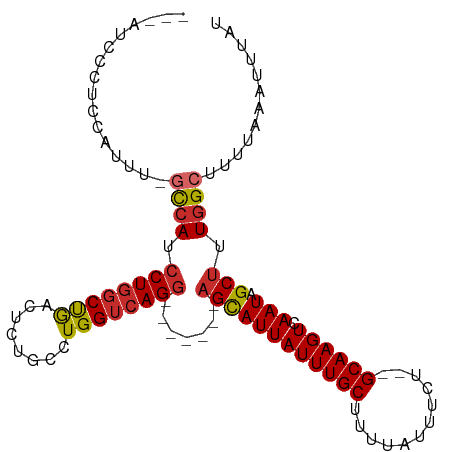
| Mean single sequence MFE | -23.81 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
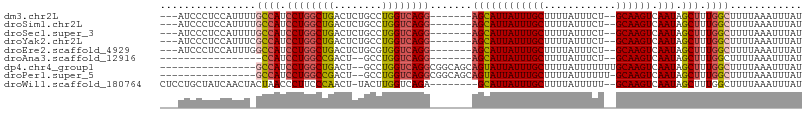
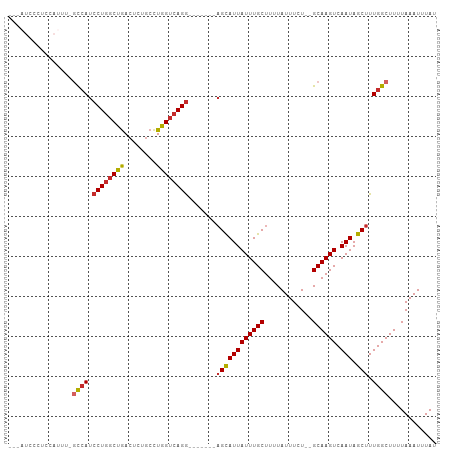
>dm3.chr2L 14754250 95 + 23011544 ---AUCCCUCCAUUUUGCCAUCCUGGCUGACUCUGCCUGGUCAGG-------AGCAUUAUUUGCUUUUAUUUCU--GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ---.............(((((((((((((........))))))))-------)(((((((((((..........--)))))).))).))..))))............ ( -23.40, z-score = -1.59, R) >droSim1.chr2L 14499977 95 + 22036055 ---AUCCCUCCAUUUUGCCAUCCUGGCUGACUCUGCCUGGUCAGG-------AGCAUUAUUUGCUUUUAUUUCU--GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ---.............(((((((((((((........))))))))-------)(((((((((((..........--)))))).))).))..))))............ ( -23.40, z-score = -1.59, R) >droSec1.super_3 1026095 95 + 7220098 ---AUCCCUCCAUUUUGCCAUCCUGGCUGACUCUGCCUGGUCAGG-------AGCAUUAUUUGCUUUUAUUUCU--GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ---.............(((((((((((((........))))))))-------)(((((((((((..........--)))))).))).))..))))............ ( -23.40, z-score = -1.59, R) >droYak2.chr2L 14859707 95 + 22324452 ---AUCCCUCCAUUUCGCCAUCCUGGCUGACUCUGCCUGGUCAGG-------AGCAUUAUUUGCUUUUAUUUCU--GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ---.............(((((((((((((........))))))))-------)(((((((((((..........--)))))).))).))..))))............ ( -23.70, z-score = -1.89, R) >droEre2.scaffold_4929 6503157 95 + 26641161 ---AUCCCUCCAUUUGGCCAUCCUGGCUGACUCUGCGUGGUCAGG-------AGCAUUAUUUGCUUUUAUUUCU--GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ---............(((((..(((..(((((.((((..(..(((-------((((.....)))))))..)..)--)))))))).)))...)))))........... ( -25.30, z-score = -1.73, R) >droAna3.scaffold_12916 15089517 79 - 16180835 -----------------CCAUCCUGGCCGACU--GCCUGGUCAGG-------AGCAUUAUUUGCUUUUAUUUCU--GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU -----------------((((((((((((...--...))))))))-------)(((((((((((..........--)))))).))).))..)))............. ( -22.30, z-score = -2.12, R) >dp4.chr4_group1 2261338 89 - 5278887 ----------------GCCAUCCUGGCUGACU--GCCUGGUCAGGCGGCAGCAGUAUUAUUUGCUUUUAUUUUUUUGCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ----------------((((.....((((.((--((((....))))))))))..((((((((((............)))))).))))....))))............ ( -27.50, z-score = -2.08, R) >droPer1.super_5 2203006 88 + 6813705 ----------------GCCAUCCUGGCCGACU--GCCUGGUCAGGCGGCAGCAGUAUUAUUUGCUUUUAUUUUUU-GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ----------------(((..((((((((...--...)))))))).)))(((((......))))).......(((-(.(((((((.....))))))).))))..... ( -28.20, z-score = -2.41, R) >droWil1.scaffold_180764 767577 96 - 3949147 CUCCUGCUAUCAACUACUAACCCUUCCCAACU-UACUUGGUCAGA--------GCAUUAUUUGCUUUUAUUUUU--GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ................................-.....(((((((--------(((((((((((..........--)))))).))).)))))))))........... ( -17.10, z-score = -1.98, R) >consensus ___AUCCCUCCAUUU_GCCAUCCUGGCUGACUCUGCCUGGUCAGG_______AGCAUUAUUUGCUUUUAUUUCU__GCAAGUCAAUAGCUUUGGCUUUUAAAUUUAU ................((((.((((((((........)))))))).......((((((((((((............)))))).))).))).))))............ (-17.08 = -17.27 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:25 2011