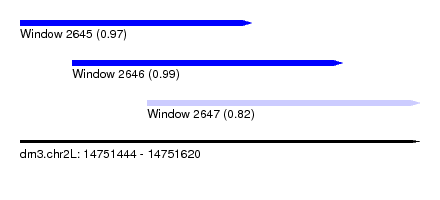
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,751,444 – 14,751,620 |
| Length | 176 |
| Max. P | 0.985451 |

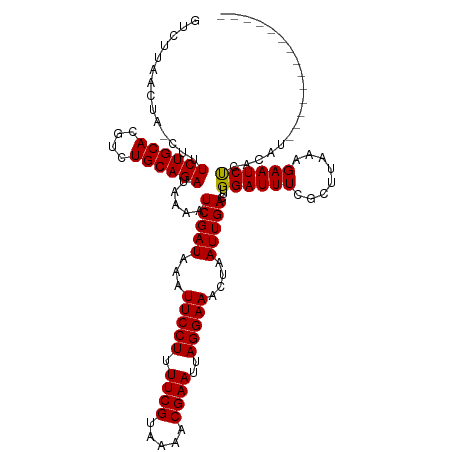
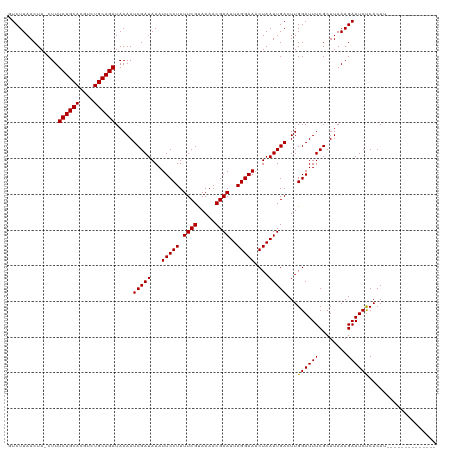
| Location | 14,751,444 – 14,751,546 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.45 |
| Shannon entropy | 0.29873 |
| G+C content | 0.35030 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -17.65 |
| Energy contribution | -16.49 |
| Covariance contribution | -1.15 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
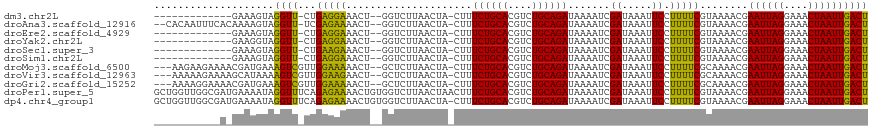
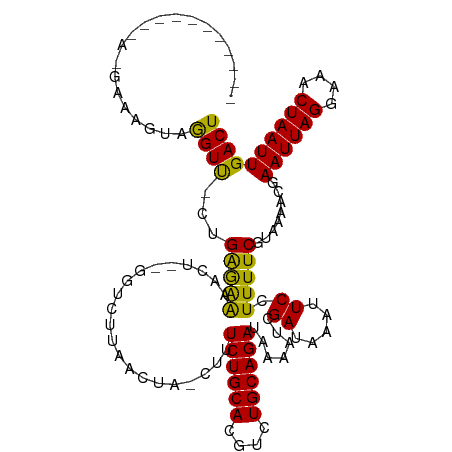
>dm3.chr2L 14751444 102 + 23011544 -------------GAAAGUAGGUU-CUGAGGAAACU--GGUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU -------------((((((((...-.(.((....))--.)......)))-)))))((((....)))).......(((((...(((((.((((.....))))..)))))....))))).. ( -25.30, z-score = -2.73, R) >droAna3.scaffold_12916 15087240 113 - 16180835 --CACAAUUUCACAAAAGUAGGUU-UCGAGAAAACU--GGUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU --..(((((((....((((((...-.(.((....))--.)......)))-)))((((((....)))))).......))....(((((.((((.....))))..)))))...)))))... ( -22.90, z-score = -1.96, R) >droEre2.scaffold_4929 6500177 102 + 26641161 -------------GAAAGUAGGUU-CUGAGGAAACU--GGUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU -------------((((((((...-.(.((....))--.)......)))-)))))((((....)))).......(((((...(((((.((((.....))))..)))))....))))).. ( -25.30, z-score = -2.73, R) >droYak2.chr2L 14856670 102 + 22324452 -------------GAAGGUAGGUU-CUGAGGAAACU--GGUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU -------------((((((((...-.(.((....))--.)......)))-)))))((((....)))).......(((((...(((((.((((.....))))..)))))....))))).. ( -25.00, z-score = -2.31, R) >droSec1.super_3 1023286 102 + 7220098 -------------GAAAGUAGGUU-CUGAAGAAACU--GGUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU -------------((((((((...-...((((....--..))))..)))-)))))((((....)))).......(((((...(((((.((((.....))))..)))))....))))).. ( -23.60, z-score = -2.47, R) >droSim1.chr2L 14497241 102 + 22036055 -------------GAAAGUAGGUU-CUGAGGAAACU--GGUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU -------------((((((((...-.(.((....))--.)......)))-)))))((((....)))).......(((((...(((((.((((.....))))..)))))....))))).. ( -25.30, z-score = -2.73, R) >droMoj3.scaffold_6500 8896677 113 + 32352404 ---AAGAAGAAAACGAUGAAAGUCGUUGGAAAAACU--GCUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACU ---..(((((.((((((....))))))((((.....--(.......)..-...((((((....)))))).............)))))))))........((((((....)))))).... ( -23.80, z-score = -2.16, R) >droVir3.scaffold_12963 5002114 113 + 20206255 ---AAAAAGAAAAGCAUAAAAGUCGUUGGAAGAACU--GCUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACU ---.....((((((.....((((.((((..((....--.))..)))).)-)))((((((....))))))................))))))........((((((....)))))).... ( -23.90, z-score = -2.25, R) >droGri2.scaffold_15252 491424 113 + 17193109 ---AAAAGGAAAACGAUGAAAGUCGUUGGAAAAACU--GCUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACU ---((((((((((((((....)))))).........--...........-...((((((....)))))).............)))))))).........((((((....)))))).... ( -27.70, z-score = -3.67, R) >droPer1.super_5 2194361 119 + 6813705 GCUGGUUGGCGAUGAAAAUAGGUUUCAGAGAAAACUGUGGUCUUAACUAACUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU ...((((((....((..((((.((((...)))).))))..))....)))))).((((((....)))))).....(((((...(((((.((((.....))))..)))))....))))).. ( -27.70, z-score = -2.05, R) >dp4.chr4_group1 2252957 118 - 5278887 GCUGGUUGGCGAUGAAAAUAGGUUUCAGAGAAAACUGUGGUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU ...((((..(..(((((.....)))))..)..))))(((((....))))-)..((((((....)))))).....(((((...(((((.((((.....))))..)))))....))))).. ( -27.80, z-score = -1.99, R) >consensus ___________A_GAAAGUAGGUU_CUGAGAAAACU__GGUCUUAACUA_CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACU ....................((((...(((((.....................((((((....)))))).......((.....)).)))))........((((((....)))))))))) (-17.65 = -16.49 + -1.15)
| Location | 14,751,467 – 14,751,586 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.34 |
| Shannon entropy | 0.13760 |
| G+C content | 0.33470 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -19.91 |
| Energy contribution | -19.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14751467 119 + 23011544 GUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAU .((((.....-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........))))))......))))......... ( -21.00, z-score = -1.80, R) >droAna3.scaffold_12916 15087274 105 - 16180835 GUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAU-------------- (((.......-...((((((....)))))).............(((((.((((.....))))..))))).......)))..((((((........)))))).....-------------- ( -20.20, z-score = -2.28, R) >droEre2.scaffold_4929 6500200 119 + 26641161 GUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAU .((((.....-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........))))))......))))......... ( -21.00, z-score = -1.80, R) >droYak2.chr2L 14856693 119 + 22324452 GUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAU .((((.....-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........))))))......))))......... ( -21.00, z-score = -1.80, R) >droSec1.super_3 1023309 119 + 7220098 GUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAU .((((.....-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........))))))......))))......... ( -21.00, z-score = -1.80, R) >droSim1.chr2L 14497264 119 + 22036055 GUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAU .((((.....-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........))))))......))))......... ( -21.00, z-score = -1.80, R) >droMoj3.scaffold_6500 8896711 105 + 32352404 CUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAU-------------- ..........-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........)))))).....-------------- ( -18.80, z-score = -2.12, R) >droVir3.scaffold_12963 5002148 105 + 20206255 CUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAU-------------- ..........-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........)))))).....-------------- ( -18.80, z-score = -2.12, R) >droGri2.scaffold_15252 491458 105 + 17193109 CUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAU-------------- ..........-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........)))))).....-------------- ( -18.80, z-score = -2.12, R) >droWil1.scaffold_180764 3175664 105 + 3949147 CUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAU-------------- ..........-...((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........)))))).....-------------- ( -19.60, z-score = -2.35, R) >droPer1.super_5 2194400 106 + 6813705 GUCUUAACUAACUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCCCACAU-------------- (((...........((((((....)))))).............(((((.((((.....))))..))))).......))).(((..(((.......)))..)))...-------------- ( -22.50, z-score = -3.14, R) >dp4.chr4_group1 2252996 105 - 5278887 GUCUUAACUA-CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCCCACAU-------------- (((.......-...((((((....)))))).............(((((.((((.....))))..))))).......))).(((..(((.......)))..)))...-------------- ( -22.50, z-score = -3.18, R) >consensus GUCUUAACUA_CUUUCUGCACGUCUGCAGAUAAAAUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAU______________ ..............((((((....)))))).....(((((...(((((.((((.....))))..)))))....)))))...((((((........))))))................... (-19.91 = -19.77 + -0.14)

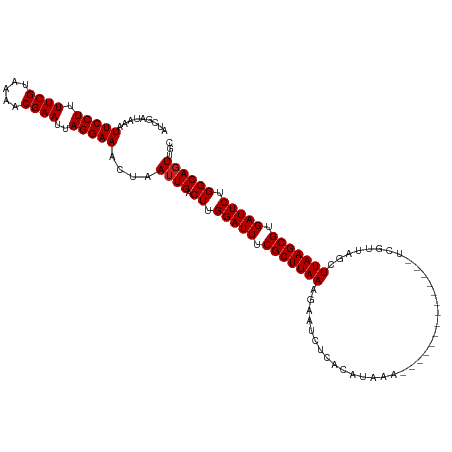
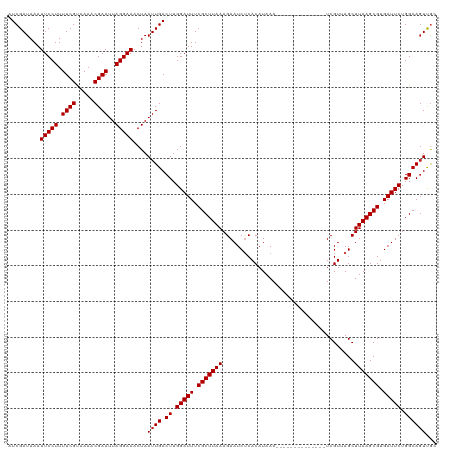
| Location | 14,751,500 – 14,751,620 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.40 |
| Shannon entropy | 0.15845 |
| G+C content | 0.35227 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14751500 120 + 23011544 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAUAAAUCGUUAGCUUAAGCGUGAUUCUGGCAGUUGC .........(((((.((((.....))))..)))))..((((((.((.(((((.((((((((((.(((.......))).)))..........(....)))))))).))))).)))))))). ( -25.80, z-score = -1.68, R) >droAna3.scaffold_12916 15087307 106 - 16180835 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAA--------------UCGUUAGCUUAAGCGUGAUUCUGGCAGUUGC .........(((((.((((.....))))..)))))..((((((.((.(((((.(((((((.....((.((.....--------------..)).)).))))))).))))).)))))))). ( -23.90, z-score = -1.67, R) >droEre2.scaffold_4929 6500233 120 + 26641161 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAUAAAUCGUUAGCUUAAGCGUGAUUCUGGCAGUUGC .........(((((.((((.....))))..)))))..((((((.((.(((((.((((((((((.(((.......))).)))..........(....)))))))).))))).)))))))). ( -25.80, z-score = -1.68, R) >droYak2.chr2L 14856726 120 + 22324452 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAUAAAUCGUUAGCUUAAGCGUGAUUCUGGCAGUUGC .........(((((.((((.....))))..)))))..((((((.((.(((((.((((((((((.(((.......))).)))..........(....)))))))).))))).)))))))). ( -25.80, z-score = -1.68, R) >droSec1.super_3 1023342 120 + 7220098 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAUAAAUCGUUAGCUUAAGCGUGAUUCUGGCAGUUGC .........(((((.((((.....))))..)))))..((((((.((.(((((.((((((((((.(((.......))).)))..........(....)))))))).))))).)))))))). ( -25.80, z-score = -1.68, R) >droSim1.chr2L 14497297 120 + 22036055 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAAGAAUCUCACAUAAAUCGUUAGCUUAAGCGUGAUUCUGGCAGUUGC .........(((((.((((.....))))..)))))..((((((.((.(((((.((((((((((.(((.......))).)))..........(....)))))))).))))).)))))))). ( -25.80, z-score = -1.68, R) >droMoj3.scaffold_6500 8896744 106 + 32352404 AUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAA--------------UCGUGAGCUUAAGCGUGAUUCUGGCAGUCAC .........(((((.((((.....))))..)))))......(((((.(((((.(((((((.....(((((.....--------------..))))).))))))).)))))....))))). ( -28.30, z-score = -3.03, R) >droVir3.scaffold_12963 5002181 106 + 20206255 AUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAA--------------UCGUGAGCUUAAGCGUGAUUCUGGCAGUCAC .........(((((.((((.....))))..)))))......(((((.(((((.(((((((.....(((((.....--------------..))))).))))))).)))))....))))). ( -28.30, z-score = -3.03, R) >droGri2.scaffold_15252 491491 106 + 17193109 AUCGAUAAAUUCCUUUUCGCAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAA--------------UCGUGAGCUUAAGCGUGAUUCUGGCAGUCGA .(((((...(((((.((((.....))))..)))))......((.((.(((((.(((((((.....(((((.....--------------..))))).))))))).))))).))))))))) ( -30.10, z-score = -3.06, R) >droWil1.scaffold_180764 3175697 106 + 3949147 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAA--------------UCGUGAGCUUAAGCGUGAUUCUGGCAUUUGC .(((((...(((((.((((.....))))..)))))....)))))((.(((((.(((((((.....(((((.....--------------..))))).))))))).))))).))....... ( -27.10, z-score = -2.79, R) >droPer1.super_5 2194434 106 + 6813705 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCCCACAUAAA--------------UCGUUAGCUUAAGCGUGAUUCUGGCAGUUGG .........(((((.((((.....))))..))))).(((((((.((.(((((.(((((((...............--------------........))))))).))))).))))))))) ( -26.00, z-score = -2.36, R) >dp4.chr4_group1 2253029 106 - 5278887 AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCCCACAUAAA--------------UCGUUAGCUUAAGCGUGAUUCUGGCAGUUGG .........(((((.((((.....))))..))))).(((((((.((.(((((.(((((((...............--------------........))))))).))))).))))))))) ( -26.00, z-score = -2.36, R) >consensus AUCGAUAAAUUCCUUUUCGUAAAACGAAUUAGGAAACUAAUUGACUUGGAUUUCGCUUAAAGAAUCUCACAUAAA______________UCGUUAGCUUAAGCGUGAUUCUGGCAGUUGC .........(((((.((((.....))))..)))))....((((.((.(((((.(((((((.....................................))))))).))))).))))))... (-20.44 = -20.52 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:24 2011