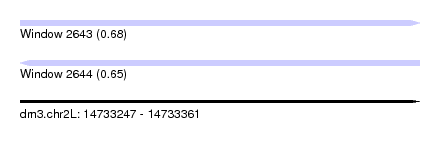
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,733,247 – 14,733,361 |
| Length | 114 |
| Max. P | 0.683304 |

| Location | 14,733,247 – 14,733,361 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 65.32 |
| Shannon entropy | 0.65101 |
| G+C content | 0.56781 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -11.47 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.57 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.683304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
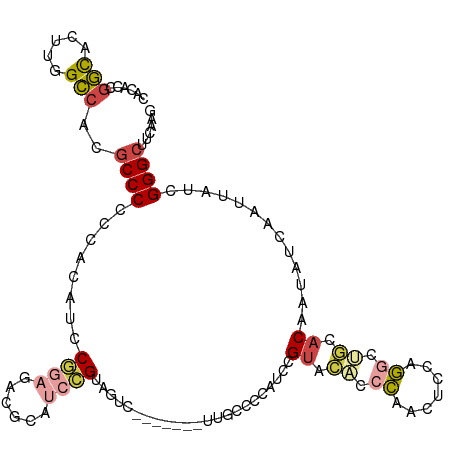
>dm3.chr2L 14733247 114 + 23011544 CACACCGGCACUUGGCCACGCCCCGCACAUCCGGAGCCGCAUCCGUAGUCCCAUCCGUUUCCCCAUCCGUACACCCAACUCCAGGCUGCACAAUAUCAAUUAUCGGGCUUCAAG ......(((.....)))..((((.........(((......)))((((((.................................))))))...............))))...... ( -20.71, z-score = 0.86, R) >droSim1.chr2L 14483348 114 + 22036055 CACACCGGCACUUUGCCACGCCCCCCACAUCCGGAGACGCAUCCGUAGUCCCAUCCGUUGCCCCAUCCGUACACCCAACUCCAGGCUGCACAAUAUCAAUUAUCGGGCUUCAAG ......(((.....)))..((((.........(((......)))((((((......((((...............))))....))))))...............))))...... ( -23.36, z-score = -0.72, R) >droSec1.super_3 1009003 113 + 7220098 CACACCGGCACUUGGCCACGCCCCC-ACAUCCGGAGACGCAUCCGUAGUCCCAUCCGUUGCCCCAUCCGUACACCCAACUCCAGGGUGCACAAUAUCAAUUAUCGGGCUUCAAG ......(((.....)))..((((..-.....(((((((((....)).)))...))))...........((.(((((.......))))).)).............))))...... ( -28.00, z-score = -1.01, R) >droYak2.chr2L 14840973 102 + 22324452 CUCACCGGUACUUGGCCACGCCCACCACAUCCGGAGCCACAUCCGUA------------UUUCUAUCCGUACACCCAACUCCGGGCCGCACAAUAUCAAUUAUCGGGCUUCAAG ....((((((.(((((...((((.........(((......)))(((------------(........))))..........)))).)).......))).))))))........ ( -21.31, z-score = -0.70, R) >droVir3.scaffold_12963 5025630 103 + 20206255 ----CAGACACUGCGUCCGCCCCCUUCUACACCCCCACCCACUGGCAAAC-------GUGCCCCGCUUGCUAGCAGCCCUUUUCCGCUUACAAUAUCAAUUAUCGGGCGUCAAG ----..(((...((((..(((......................)))..))-------))((((.(((....)))(((........)))................)))))))... ( -21.25, z-score = -1.23, R) >droMoj3.scaffold_6500 8911723 86 + 32352404 ----CAAGUAUAU-GCCCCGCCCCUUACACCC--------ACUCGC------------UAUUCGCUCUGUAAACCCUUUUGCUGU---UACGAUAUCAAUUAUCGGGCGUCAAG ----.........-....(((((.........--------....((------------.....))...(((((....)))))...---...((((.....)))))))))..... ( -13.50, z-score = -0.62, R) >consensus CACACCGGCACUUGGCCACGCCCCCCACAUCCGGAGACGCAUCCGUAGUC_______UUGCCCCAUCCGUACACCCAACUCCAGGCUGCACAAUAUCAAUUAUCGGGCUUCAAG ......(((.....)))..((((........((((......)))).......................((.((.((.......)).)).)).............))))...... (-11.47 = -12.25 + 0.78)

| Location | 14,733,247 – 14,733,361 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 65.32 |
| Shannon entropy | 0.65101 |
| G+C content | 0.56781 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.33 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.34 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
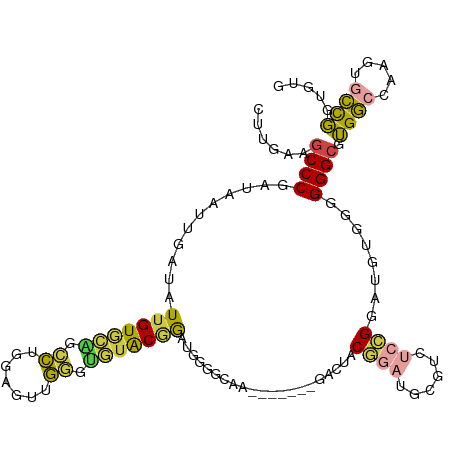
>dm3.chr2L 14733247 114 - 23011544 CUUGAAGCCCGAUAAUUGAUAUUGUGCAGCCUGGAGUUGGGUGUACGGAUGGGGAAACGGAUGGGACUACGGAUGCGGCUCCGGAUGUGCGGGGCGUGGCCAAGUGCCGGUGUG .......(((.((........(((((((.((.......)).))))))))).)))..(((..(((.(((..((..(((.(((((......))))))))..)).))).))).))). ( -35.30, z-score = 0.64, R) >droSim1.chr2L 14483348 114 - 22036055 CUUGAAGCCCGAUAAUUGAUAUUGUGCAGCCUGGAGUUGGGUGUACGGAUGGGGCAACGGAUGGGACUACGGAUGCGUCUCCGGAUGUGGGGGGCGUGGCAAAGUGCCGGUGUG ......((((.((........(((((((.((.......)).))))))))).)))).(((..(((.(((...(..((((((((.......))))))))..)..))).))).))). ( -38.80, z-score = -0.71, R) >droSec1.super_3 1009003 113 - 7220098 CUUGAAGCCCGAUAAUUGAUAUUGUGCACCCUGGAGUUGGGUGUACGGAUGGGGCAACGGAUGGGACUACGGAUGCGUCUCCGGAUGU-GGGGGCGUGGCCAAGUGCCGGUGUG ......((((.((........((((((((((.......)))))))))))).)))).(((..(((.(((..((..(((((((((....)-))))))))..)).))).))).))). ( -45.90, z-score = -2.23, R) >droYak2.chr2L 14840973 102 - 22324452 CUUGAAGCCCGAUAAUUGAUAUUGUGCGGCCCGGAGUUGGGUGUACGGAUAGAAA------------UACGGAUGUGGCUCCGGAUGUGGUGGGCGUGGCCAAGUACCGGUGAG ......((((.(((((....)))))((.(((((((((((.((...((........------------..)).)).)))))))))..)).)))))).(.(((.......))).). ( -29.10, z-score = 0.40, R) >droVir3.scaffold_12963 5025630 103 - 20206255 CUUGACGCCCGAUAAUUGAUAUUGUAAGCGGAAAAGGGCUGCUAGCAAGCGGGGCAC-------GUUUGCCAGUGGGUGGGGGUGUAGAAGGGGGCGGACGCAGUGUCUG---- .....(((((.....(((...((((.(((((.......))))).)))).))).((((-------.((..((....))..)).))))......)))))(((.....)))..---- ( -34.00, z-score = -0.90, R) >droMoj3.scaffold_6500 8911723 86 - 32352404 CUUGACGCCCGAUAAUUGAUAUCGUA---ACAGCAAAAGGGUUUACAGAGCGAAUA------------GCGAGU--------GGGUGUAAGGGGCGGGGC-AUAUACUUG---- ......((((((((.....))))...---...((....(.(..(((...((.....------------))..))--------)..).).....)).))))-.........---- ( -16.90, z-score = 0.77, R) >consensus CUUGAAGCCCGAUAAUUGAUAUUGUGCAGCCUGGAGUUGGGUGUACGGAUGGGGCAA_______GACUACGGAUGCGUCUCCGGAUGUGGGGGGCGUGGCCAAGUGCCGGUGUG ......((((...........(((((((.((.......)).))))))).....................((((......))))........)))).((((.....))))..... (-19.74 = -19.33 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:21 2011