| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,714,915 – 14,715,013 |
| Length | 98 |
| Max. P | 0.612477 |

| Location | 14,714,915 – 14,715,013 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Shannon entropy | 0.40523 |
| G+C content | 0.35671 |
| Mean single sequence MFE | -16.52 |
| Consensus MFE | -10.76 |
| Energy contribution | -10.74 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
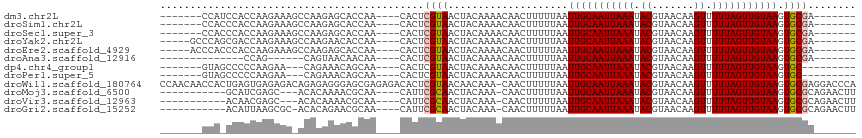
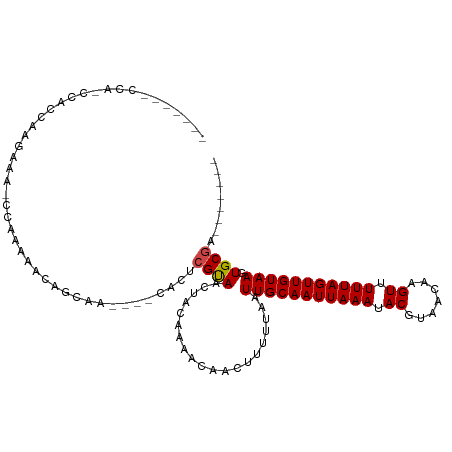
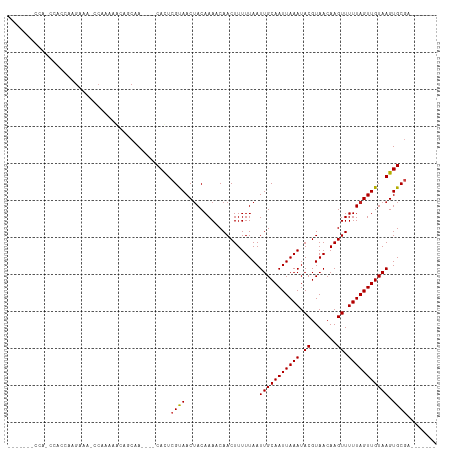
>dm3.chr2L 14714915 98 + 23011544 -------CCAUCCACCAAGAAAGCCAAGAGCACCAA----CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGA------- -------...(((((..((((((....(((......----..)))((....)).......))))))..(((((((((((.((.......)).)))))))))))))).))------- ( -16.10, z-score = -1.88, R) >droSim1.chr2L 14464436 98 + 22036055 -------CCACCCACCAAGAAAGCCAAGAGCACCAA----CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGA------- -------...(.(((..((((((....(((......----..)))((....)).......))))))..(((((((((((.((.......)).)))))))))))))).).------- ( -15.50, z-score = -1.60, R) >droSec1.super_3 990881 98 + 7220098 -------CCACCCACCAAGAAAGCCAAGAGCACCAA----CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGA------- -------...(.(((..((((((....(((......----..)))((....)).......))))))..(((((((((((.((.......)).)))))))))))))).).------- ( -15.50, z-score = -1.60, R) >droYak2.chr2L 14822036 100 + 22324452 -----GCCCAGCGACCAAGAAAGCCAAGAACACCAA----CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGA------- -----.....(((....((((((....((.......----...))((....)).......))))))..(((((((((((.((.......)).))))))))))).)))..------- ( -12.70, z-score = 0.12, R) >droEre2.scaffold_4929 6464128 100 + 26641161 -----ACCCACCCACCAAGAAAGCCAAGAGCACCAA----CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGA------- -----.....(.(((..((((((....(((......----..)))((....)).......))))))..(((((((((((.((.......)).)))))))))))))).).------- ( -15.50, z-score = -1.72, R) >droAna3.scaffold_12916 15058063 85 - 16180835 --------------CCAG------CAGUAACAACAA----CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGA------- --------------...(------(((((...((..----.....))...)))...............(((((((((((.((.......)).))))))))))).)))..------- ( -13.20, z-score = -0.51, R) >dp4.chr4_group1 2187115 93 - 5278887 -------GUAGCCCCCAAGAA---CAGAAACAGCAA----CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGC--------- -------..............---............----((((..((((((.(((((.((..((((((....))))))...)).....)))))))))))..)))).--------- ( -11.90, z-score = 0.01, R) >droPer1.super_5 2129337 93 + 6813705 -------GUAGCCCCCAAGAA---CAGAAACAGCAA----CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGC--------- -------..............---............----((((..((((((.(((((.((..((((((....))))))...)).....)))))))))))..)))).--------- ( -11.90, z-score = 0.01, R) >droWil1.scaffold_180764 822431 115 - 3949147 CCAACAACCACUGAGUGAGAGACAGAGAGGGAGCGAGAGACACUCGUAACAACAAA-CAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGAGGACCCA ..........(((..(....).)))...(((.(((((.....))))).........-...((((.((.(((((((((((.((.......)).))))))))))).)).)))).))). ( -24.70, z-score = -1.76, R) >droMoj3.scaffold_6500 23421565 97 - 32352404 -----------GCAUCGAGC---ACACAAAACGCAA----CAUUCGCAACUACAAA-CAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGCAGAACUU -----------...((..((---((.......((..----.....)).........-...........(((((((((((.((.......)).)))))))))))))))...)).... ( -19.00, z-score = -1.62, R) >droVir3.scaffold_12963 15163177 97 - 20206255 -----------ACAACGAGC---ACACAAAACGCAA----CAUUCGCAACUACAAA-CAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGCAGAACUU -----------.......((---.(((.....((..----.....)).........-...........(((((((((((.((.......)).)))))))))))))).))....... ( -18.40, z-score = -1.69, R) >droGri2.scaffold_15252 453426 99 + 17193109 -----------ACAUUAAGCGC-ACACAGAACGCAA----CAUUCGCAACUACAAA-CAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGCAGAACUU -----------.......((((-((.......((..----.....)).........-...........(((((((((((.((.......)).)))))))))))))))))....... ( -23.80, z-score = -3.24, R) >consensus _______CCA_CCACCAAGAAA_CCAAAAACAGCAA____CACUCGUAACUACAAAACAACUUUUUAAUUGCAAUUAAAUACGUAACAAGUUUUUAGUUGUAAGUGCGA_______ ............................................((((....................(((((((((((.((.......)).))))))))))).))))........ (-10.76 = -10.74 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:19 2011