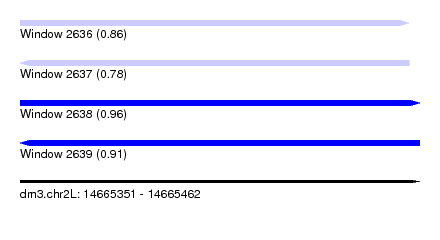
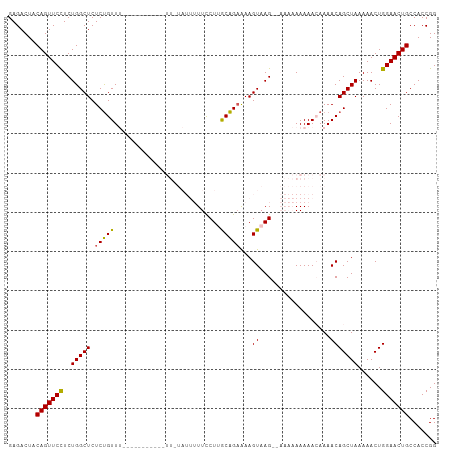
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,665,351 – 14,665,462 |
| Length | 111 |
| Max. P | 0.956948 |

| Location | 14,665,351 – 14,665,459 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.24 |
| Shannon entropy | 0.27799 |
| G+C content | 0.39359 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -15.99 |
| Energy contribution | -15.71 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.863826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14665351 108 + 23011544 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUGUUUUUUUUUU-CUUACUUUUCUGUAAGGAAAAAUAAAACAGAAAAAAAAAAACAGAGAGCCAGAGGAACUGUAGUCUC ..((..((((((((.((((((.....(((((((((((((((-(((((......))))))))))).)))))))))....)))))).(.....)...))))))))..)).. ( -33.40, z-score = -3.57, R) >droEre2.scaffold_4929 6412606 83 + 26641161 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUGUUUUU------CUUACUUUUCCGCCAGGAA--------------------AACGGAGAGCCAGAAGAACUGUAGUCUC ..((..(((((((((((.....))))(((((.((((------((...((((((....))))--------------------)).)))))).))))))))))))..)).. ( -25.30, z-score = -2.08, R) >droYak2.chr2L 14771431 83 + 22324452 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUGUUUUU------CUUACUUCACUGCCAGGAA--------------------AACAGAGAGCCAGAAGAACUGUAGUCUC ..((..(((((((....((((.(((.((((((((((------((..(......)..)))))--------------------)))))))))).)))))))))))..)).. ( -23.80, z-score = -1.51, R) >droSec1.super_3 938869 96 + 7220098 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUUUUUUUUUUU--CUUACUUUUCUGCAAGAAAAAAUACAA-----------AAACAGAGAGCCAGAGGAACUGUAGUCUC ..((..((((((((.((((....((((((((...((((((--(((.(......).)))))))))...))-----------)))))).))))....))))))))..)).. ( -28.30, z-score = -3.02, R) >droSim1.chr2L 14414144 98 + 22036055 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUUUUUUUUUUUUUCUUACUUUUCUGCAAGAAAAAAUACAA-----------AAACAGAGAGCCAGAGGAACUGUAGUCUC ..((..((((((((.((((....((((((((.....(((((((((.(......).)))))))))...))-----------)))))).))))....))))))))..)).. ( -27.00, z-score = -2.53, R) >consensus CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUGUUUUUUUUU__CUUACUUUUCUGCAAGGAAAAAUA_AA___________AAACAGAGAGCCAGAGGAACUGUAGUCUC ..((..(((((((((((.....)))).....................(((((((.............................)))))))......)))))))..)).. (-15.99 = -15.71 + -0.28)

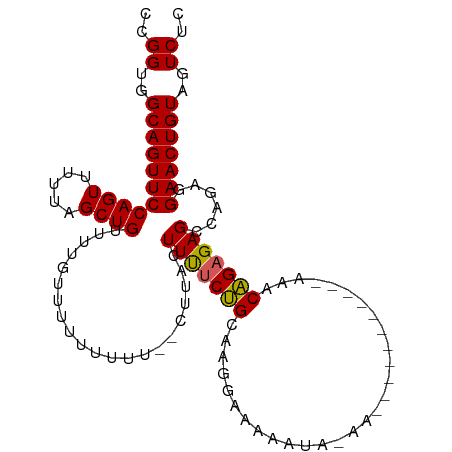
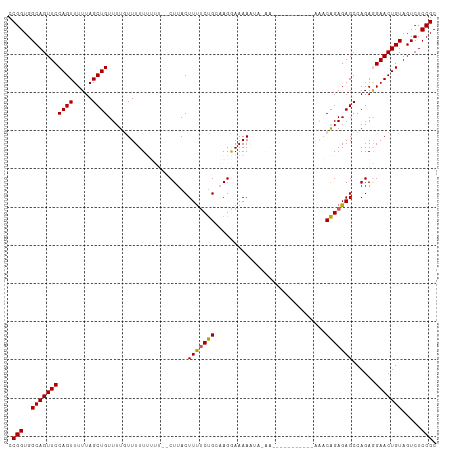
| Location | 14,665,351 – 14,665,459 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Shannon entropy | 0.27799 |
| G+C content | 0.39359 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -12.96 |
| Energy contribution | -12.44 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14665351 108 - 23011544 GAGACUACAGUUCCUCUGGCUCUCUGUUUUUUUUUUUCUGUUUUAUUUUUCCUUACAGAAAAGUAAG-AAAAAAAAAACAAAACAGCUAAAAACUGGAACUGCCACCGG .......(((((((.........(((((((.((((((...(((((((((((......))))))))))-)...)))))).))))))).........)))))))....... ( -27.87, z-score = -3.30, R) >droEre2.scaffold_4929 6412606 83 - 26641161 GAGACUACAGUUCUUCUGGCUCUCCGUU--------------------UUCCUGGCGGAAAAGUAAG------AAAAACAAAACAGCUAAAAACUGGAACUGCCACCGG .......((((((((((.(((.((((((--------------------.....))))))..))).))------))........(((.......)))))))))....... ( -19.60, z-score = -1.09, R) >droYak2.chr2L 14771431 83 - 22324452 GAGACUACAGUUCUUCUGGCUCUCUGUU--------------------UUCCUGGCAGUGAAGUAAG------AAAAACAAAACAGCUAAAAACUGGAACUGCCACCGG .......((((((((((.((((.(((((--------------------.....))))).).))).))------))........(((.......)))))))))....... ( -18.40, z-score = -0.42, R) >droSec1.super_3 938869 96 - 7220098 GAGACUACAGUUCCUCUGGCUCUCUGUUU-----------UUGUAUUUUUUCUUGCAGAAAAGUAAG--AAAAAAAAAAAAAACAGCUAAAAACUGGAACUGCCACCGG .......(((((((.........((((((-----------((...(((((((((((......)))))--))))))...)))))))).........)))))))....... ( -28.67, z-score = -4.25, R) >droSim1.chr2L 14414144 98 - 22036055 GAGACUACAGUUCCUCUGGCUCUCUGUUU-----------UUGUAUUUUUUCUUGCAGAAAAGUAAGAAAAAAAAAAAAAAAACAGCUAAAAACUGGAACUGCCACCGG .......(((((((.........((((((-----------((.(.(((((((((((......))))))))))).)...)))))))).........)))))))....... ( -27.37, z-score = -3.79, R) >consensus GAGACUACAGUUCCUCUGGCUCUCUGUUU___________UU_UAUUUUUCCUUGCAGAAAAGUAAG__AAAAAAAAACAAAACAGCUAAAAACUGGAACUGCCACCGG .......(((((((..(((((.(((((...........................)))))...((..................)))))))......)))))))....... (-12.96 = -12.44 + -0.52)

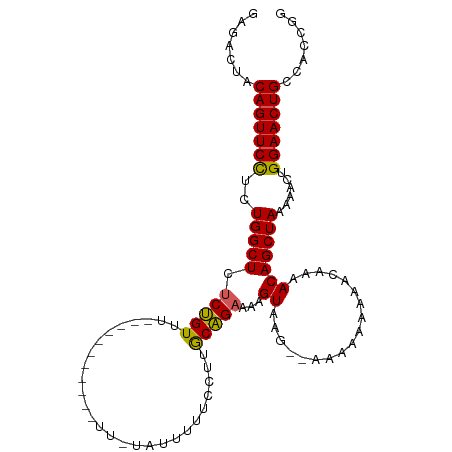
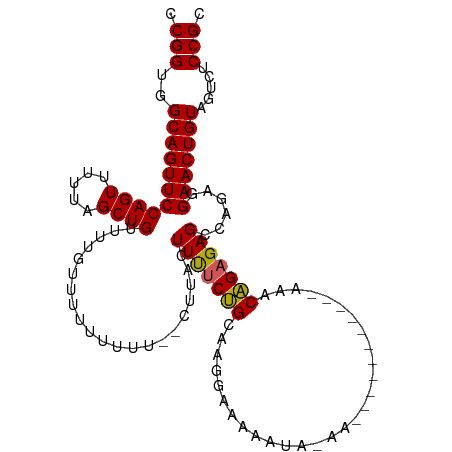
| Location | 14,665,351 – 14,665,462 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.75 |
| Shannon entropy | 0.27054 |
| G+C content | 0.41246 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -17.73 |
| Energy contribution | -17.45 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.956948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14665351 111 + 23011544 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUGUUUUUUUUUU-CUUACUUUUCUGUAAGGAAAAAUAAAACAGAAAAAAAAAAACAGAGAGCCAGAGGAACUGUAGUCUCCGC .(((..((((((((.((((((.....(((((((((((((((-(((((......))))))))))).)))))))))....)))))).(.....)...)))))))).....))). ( -35.30, z-score = -3.82, R) >droEre2.scaffold_4929 6412606 86 + 26641161 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUGUUUUU------CUUACUUUUCCGCCAGGAAAA--------------------CGGAGAGCCAGAAGAACUGUAGUCUCCGC .(((..(((((((((((.....))))(((((.((((------((...((((((....))))))--------------------.)))))).)))))))))))).....))). ( -27.20, z-score = -2.26, R) >droYak2.chr2L 14771431 86 + 22324452 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUGUUUUU------CUUACUUCACUGCCAGGAAAA--------------------CAGAGAGCCAGAAGAACUGUAGUCUCCGC .(((.(((.....((((((((.(((.((((((((((------((..(......)..)))))))--------------------))))))))...))))))))..))).))). ( -26.90, z-score = -2.23, R) >droSec1.super_3 938869 99 + 7220098 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUUUUUUUUUUU--CUUACUUUUCUGCAAGAAAAAAUACAA-----------AAACAGAGAGCCAGAGGAACUGUAGUCUCCGC .(((..((((((((.((((....((((((((...((((((--(((.(......).)))))))))...))-----------)))))).))))....)))))))).....))). ( -30.20, z-score = -3.25, R) >droSim1.chr2L 14414144 101 + 22036055 CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUUUUUUUUUUUUUCUUACUUUUCUGCAAGAAAAAAUACAA-----------AAACAGAGAGCCAGAGGAACUGUAGUCUCCGC .(((..((((((((.((((....((((((((.....(((((((((.(......).)))))))))...))-----------)))))).))))....)))))))).....))). ( -28.90, z-score = -2.80, R) >consensus CCGGUGGCAGUUCCAGUUUUUAGCUGUUUUGUUUUUUUUU__CUUACUUUUCUGCAAGGAAAAAUA_AA___________AAACAGAGAGCCAGAGGAACUGUAGUCUCCGC .(((..(((((((((((.....)))).....................(((((((.............................)))))))......))))))).....))). (-17.73 = -17.45 + -0.28)

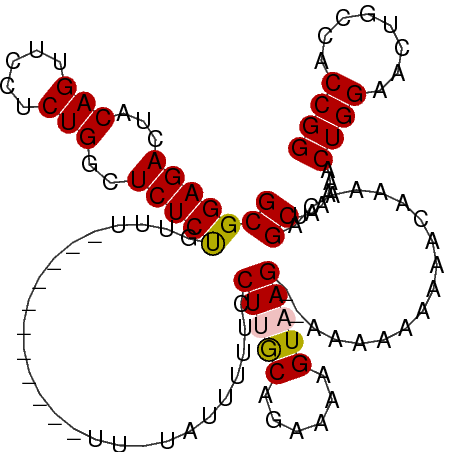
| Location | 14,665,351 – 14,665,462 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Shannon entropy | 0.27054 |
| G+C content | 0.41246 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14665351 111 - 23011544 GCGGAGACUACAGUUCCUCUGGCUCUCUGUUUUUUUUUUUCUGUUUUAUUUUUCCUUACAGAAAAGUAAG-AAAAAAAAAACAAAACAGCUAAAAACUGGAACUGCCACCGG .(((.(....(((((((.........(((((((.((((((...(((((((((((......))))))))))-)...)))))).))))))).........))))))).).))). ( -30.47, z-score = -3.50, R) >droEre2.scaffold_4929 6412606 86 - 26641161 GCGGAGACUACAGUUCUUCUGGCUCUCCG--------------------UUUUCCUGGCGGAAAAGUAAG------AAAAACAAAACAGCUAAAAACUGGAACUGCCACCGG (((((((...(((.....)))..))))))--------------------).....((((((....((...------....))....(((.......)))...)))))).... ( -22.20, z-score = -1.23, R) >droYak2.chr2L 14771431 86 - 22324452 GCGGAGACUACAGUUCUUCUGGCUCUCUG--------------------UUUUCCUGGCAGUGAAGUAAG------AAAAACAAAACAGCUAAAAACUGGAACUGCCACCGG (((((((...(((.....)))..))))))--------------------).....(((((((..(((...------...................)))...))))))).... ( -21.95, z-score = -0.96, R) >droSec1.super_3 938869 99 - 7220098 GCGGAGACUACAGUUCCUCUGGCUCUCUGUUU-----------UUGUAUUUUUUCUUGCAGAAAAGUAAG--AAAAAAAAAAAAAACAGCUAAAAACUGGAACUGCCACCGG .(((.(....(((((((.........((((((-----------((...(((((((((((......)))))--))))))...)))))))).........))))))).).))). ( -31.27, z-score = -4.31, R) >droSim1.chr2L 14414144 101 - 22036055 GCGGAGACUACAGUUCCUCUGGCUCUCUGUUU-----------UUGUAUUUUUUCUUGCAGAAAAGUAAGAAAAAAAAAAAAAAAACAGCUAAAAACUGGAACUGCCACCGG .(((.(....(((((((.........((((((-----------((.(.(((((((((((......))))))))))).)...)))))))).........))))))).).))). ( -29.97, z-score = -3.93, R) >consensus GCGGAGACUACAGUUCCUCUGGCUCUCUGUUU___________UU_UAUUUUUCCUUGCAGAAAAGUAAG__AAAAAAAAACAAAACAGCUAAAAACUGGAACUGCCACCGG (((((((...(((.....)))..)))))..........................(((((......)))))..................))......((((........)))) (-15.28 = -15.36 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:17 2011