| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,665,248 – 14,665,341 |
| Length | 93 |
| Max. P | 0.993161 |

| Location | 14,665,248 – 14,665,341 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.41888 |
| G+C content | 0.48260 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.02 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
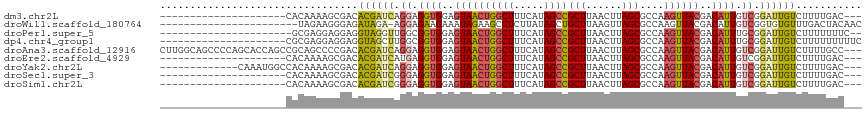
>dm3.chr2L 14665248 93 - 23011544 ---------------------CACAAAAGCGACACGAUCAGGAGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUGUCGGAUUGUCUUUUGAC--- ---------------------..((((((....((((((.((..(((..((((((((((.....))))(((......)))....))))))..)))..)).))))))))))))..--- ( -31.80, z-score = -3.56, R) >droWil1.scaffold_180764 911710 93 + 3949147 -----------------------UAGAAGGGAGAUAGA-AGGAGAAGAAAUAGAAGCCUCUUAUAGCUGCUUAAGUUAGCGCCAAGUUACGACAUUGUCGGUGUGUUUGACUACAAC -----------------------....(((.((.((..-(((((.............))))).)).)).))).(((((((((.......((((...))))..))).))))))..... ( -13.72, z-score = 1.71, R) >droPer1.super_5 2061633 93 - 6813705 ----------------------GCGAGGAGGAGGUAGGUUGGCGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUUGCGGAUUGUCUUUUUUUC-- ----------------------..(((((((((..((.((.((((((..((((((((((.....))))(((......)))....))))))..)).)))).))))..)))))))))-- ( -32.20, z-score = -2.45, R) >dp4.chr4_group1 2116180 96 + 5278887 ---------------------CGCGAGGAGGAGGUAGCUUGGCGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUUGCGGAUUGUCUUUUUUUUUC ---------------------...((((((((((((((((((((.(((..(((.(((((.....))))).)))..))).)))))))))))((((.........)))).))))))))) ( -36.40, z-score = -3.44, R) >droAna3.scaffold_12916 15011151 114 + 16180835 CUUGGCAGCCCCAGCACCAGCCGCAGCCCCGACACGAUCAGGAGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUGUCGGAUUGUCUUUUGCC--- ...((((((..(.((....)).)..))......((((((.((..(((..((((((((((.....))))(((......)))....))))))..)))..)).))))))....))))--- ( -34.30, z-score = -0.49, R) >droEre2.scaffold_4929 6412503 93 - 26641161 ---------------------CACAAAAGCGACACGAUCAUGAGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUGUCGGAUUGUCUUUUGAC--- ---------------------..(((((((((..((((.....((((..((((((((((.....))))(((......)))....))))))..))))))))..))).))))))..--- ( -31.00, z-score = -3.48, R) >droYak2.chr2L 14771320 101 - 22324452 -------------CAAAUGGCCACAAAAGCGACACGAUCAGGAGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUGUCGGAUUGUCUUUUGAC--- -------------..........((((((....((((((.((..(((..((((((((((.....))))(((......)))....))))))..)))..)).))))))))))))..--- ( -31.80, z-score = -2.61, R) >droSec1.super_3 938766 93 - 7220098 ---------------------CACAAAAGCGACACGAUCGGGAGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUGUCGGAUUGUCUUUUGAC--- ---------------------..((((((....((((((.((..(((..((((((((((.....))))(((......)))....))))))..)))..)).))))))))))))..--- ( -33.60, z-score = -3.89, R) >droSim1.chr2L 14414041 93 - 22036055 ---------------------CACAAAAGCGACACGAUCGGGAGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUGUCGGAUUGUCUUUUGAC--- ---------------------..((((((....((((((.((..(((..((((((((((.....))))(((......)))....))))))..)))..)).))))))))))))..--- ( -33.60, z-score = -3.89, R) >consensus _____________________CACAAAAGCGACACGAUCAGGAGGUGGAGUAACUGGCUUUCAUAGCCGCUUAACUUAGCGCCAAGUUACGACAUUGUCGGAUUGUCUUUUGAC___ .................................((((((.((.((((..((((((((((.....))))(((......)))....))))))..)))).)).))))))........... (-21.04 = -21.02 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:14 2011