| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,663,386 – 14,663,452 |
| Length | 66 |
| Max. P | 0.999066 |

| Location | 14,663,386 – 14,663,452 |
|---|---|
| Length | 66 |
| Sequences | 11 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 63.29 |
| Shannon entropy | 0.67473 |
| G+C content | 0.41574 |
| Mean single sequence MFE | -15.26 |
| Consensus MFE | -8.38 |
| Energy contribution | -8.33 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
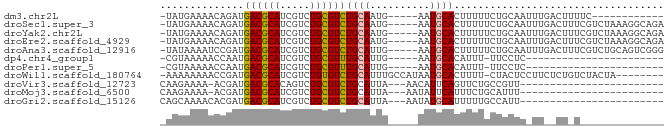
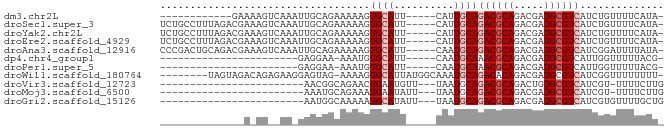
>dm3.chr2L 14663386 66 + 23011544 -UAUGAAAACAGAUGACGCAUCGUCUGCGUCUGCAAUG-----AAUGCACUUUUUCUGCAAUUUGACUUUUC------------ -...((((((((((((((((.....))))))((((..(-----((......)))..)))))))))..)))))------------ ( -15.30, z-score = -1.55, R) >droSec1.super_3 936931 78 + 7220098 -UAUGAAAACAGAUGACGCAUCGUCUGCGUCUGCAAUG-----AAUGCACUUUUUCUGCAAUUUGACUUUCGUCUAAAGGCAGA -.............((((((.....))))))((((...-----..)))).....(((((.....(((....))).....))))) ( -20.20, z-score = -1.38, R) >droYak2.chr2L 14769420 78 + 22324452 -UAUGAAAACAGAUGACGCAUCGUCUGCGUCUGCAAUG-----AAUGCACUUUUUCUGCAAUUUGACUUUCGUCUAAAGGCAGA -.............((((((.....))))))((((...-----..)))).....(((((.....(((....))).....))))) ( -20.20, z-score = -1.38, R) >droEre2.scaffold_4929 6410619 78 + 26641161 -UAUGAAAACAGAUGACGCAUCGUCUGCGUCUGCAAUG-----AAUGCACUUUUUCUGCAAUUUGACUUUCGUCUAAAGGCAGA -.............((((((.....))))))((((...-----..)))).....(((((.....(((....))).....))))) ( -20.20, z-score = -1.38, R) >droAna3.scaffold_12916 15009461 78 - 16180835 -UAUAAAAUCCGAUGACGCAUCGUCUGCGUCUGCAUUG-----AAUGCACUUUUUCUGCAAUUUGACUUUCGUCUGCAGUCGGG -........(((((((((((.....))))))((((...-----..((((.......))))....(((....)))))))))))). ( -22.90, z-score = -2.54, R) >dp4.chr4_group1 2113198 54 - 5278887 -CGUAAAAACCAAUGACGCAUCGUCUGCGUUUGCAUUG-----AAUGCACAUUU-UUCCUC----------------------- -.........((((((((((.....)))))...)))))-----...........-......----------------------- ( -8.70, z-score = -0.43, R) >droPer1.super_5 2058620 54 + 6813705 -CGUAAAAACCAAUGACGCAUCGUCUGCGUUUGCAUUG-----AAUGCACAUUU-UUCCUC----------------------- -.........((((((((((.....)))))...)))))-----...........-......----------------------- ( -8.70, z-score = -0.43, R) >droWil1.scaffold_180764 3035054 74 + 3949147 -AAAAAAAACCGAUGACGCAUCGUCUGUGUCUGCAUUUGCCAUAAUGCACUUUU-CUACUCCUUCUCUGUCUACUA-------- -..........(((((((((.....))))))((((((......)))))).....-.............))).....-------- ( -12.70, z-score = -3.21, R) >droVir3.scaffold_12723 2539167 56 + 5802038 CAAGAAAA-ACGAUGACGCACAGUCUGCGUCUGCAUUA---AACAUUCAGUUCUGCCGUU------------------------ .......(-(((..((((((.....)))))).(((...---(((.....))).)))))))------------------------ ( -11.60, z-score = -0.69, R) >droMoj3.scaffold_6500 15735223 56 + 32352404 CAAGAAAA-ACGAUGACGCAUCGUCUGCGUCUGCAUUA---AAUAUUCAUUUCUGCAUUU------------------------ ........-.....((((((.....))))))((((..(---(((....)))).))))...------------------------ ( -10.10, z-score = -0.76, R) >droGri2.scaffold_15126 987435 57 - 8399593 CAGCAAAACACGAUGACGCAUCGUCUGCGUCUGCAUUA---AAUAUGCAUUUUUGCCAUU------------------------ ..((((((......((((((.....))))))(((((..---...))))).))))))....------------------------ ( -17.30, z-score = -2.90, R) >consensus _UAUAAAAACCGAUGACGCAUCGUCUGCGUCUGCAUUG_____AAUGCACUUUUUCUGCUA_UU__CU_U______________ ..............((((((.....))))))((((..........))))................................... ( -8.38 = -8.33 + -0.05)
| Location | 14,663,386 – 14,663,452 |
|---|---|
| Length | 66 |
| Sequences | 11 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 63.29 |
| Shannon entropy | 0.67473 |
| G+C content | 0.41574 |
| Mean single sequence MFE | -19.94 |
| Consensus MFE | -9.16 |
| Energy contribution | -9.62 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
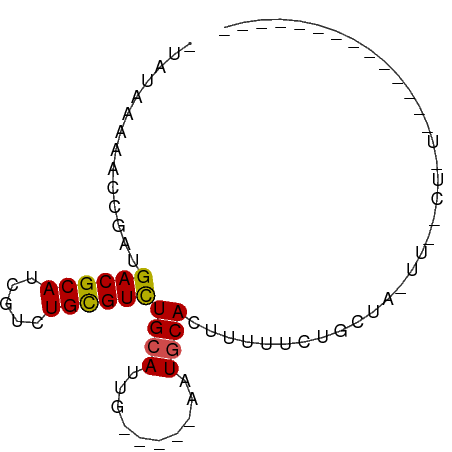
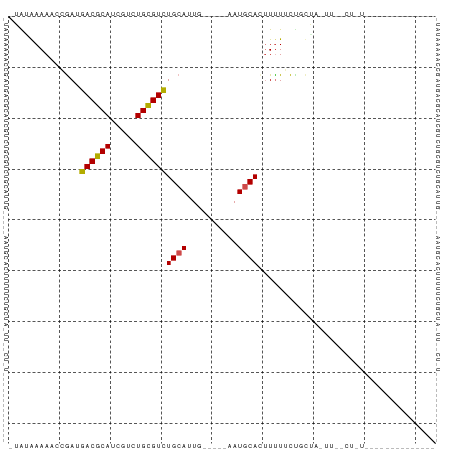
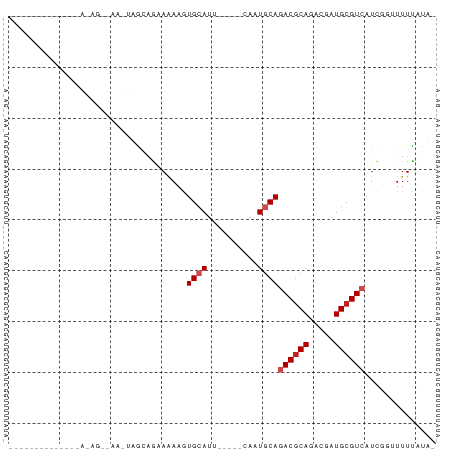
>dm3.chr2L 14663386 66 - 23011544 ------------GAAAAGUCAAAUUGCAGAAAAAGUGCAUU-----CAUUGCAGACGCAGACGAUGCGUCAUCUGUUUUCAUA- ------------((((.........(((((.....((((..-----...))))((((((.....)))))).)))))))))...- ( -18.70, z-score = -2.18, R) >droSec1.super_3 936931 78 - 7220098 UCUGCCUUUAGACGAAAGUCAAAUUGCAGAAAAAGUGCAUU-----CAUUGCAGACGCAGACGAUGCGUCAUCUGUUUUCAUA- (((((..((.(((....))).))..))))).....((((..-----...))))((((((.....)))))).............- ( -26.80, z-score = -3.73, R) >droYak2.chr2L 14769420 78 - 22324452 UCUGCCUUUAGACGAAAGUCAAAUUGCAGAAAAAGUGCAUU-----CAUUGCAGACGCAGACGAUGCGUCAUCUGUUUUCAUA- (((((..((.(((....))).))..))))).....((((..-----...))))((((((.....)))))).............- ( -26.80, z-score = -3.73, R) >droEre2.scaffold_4929 6410619 78 - 26641161 UCUGCCUUUAGACGAAAGUCAAAUUGCAGAAAAAGUGCAUU-----CAUUGCAGACGCAGACGAUGCGUCAUCUGUUUUCAUA- (((((..((.(((....))).))..))))).....((((..-----...))))((((((.....)))))).............- ( -26.80, z-score = -3.73, R) >droAna3.scaffold_12916 15009461 78 + 16180835 CCCGACUGCAGACGAAAGUCAAAUUGCAGAAAAAGUGCAUU-----CAAUGCAGACGCAGACGAUGCGUCAUCGGAUUUUAUA- .((((((((((((....)))....)))))......(((((.-----..)))))((((((.....)))))).))))........- ( -29.10, z-score = -4.90, R) >dp4.chr4_group1 2113198 54 + 5278887 -----------------------GAGGAA-AAAUGUGCAUU-----CAAUGCAAACGCAGACGAUGCGUCAUUGGUUUUUACG- -----------------------(((((.-.((((((((((-----...(((....)))...)))))).))))..)))))...- ( -14.60, z-score = -1.87, R) >droPer1.super_5 2058620 54 - 6813705 -----------------------GAGGAA-AAAUGUGCAUU-----CAAUGCAAACGCAGACGAUGCGUCAUUGGUUUUUACG- -----------------------(((((.-.((((((((((-----...(((....)))...)))))).))))..)))))...- ( -14.60, z-score = -1.87, R) >droWil1.scaffold_180764 3035054 74 - 3949147 --------UAGUAGACAGAGAAGGAGUAG-AAAAGUGCAUUAUGGCAAAUGCAGACACAGACGAUGCGUCAUCGGUUUUUUUU- --------..........((((((((..(-(....((((((......))))))(((.((.....)).))).))..))))))))- ( -13.40, z-score = -0.94, R) >droVir3.scaffold_12723 2539167 56 - 5802038 ------------------------AACGGCAGAACUGAAUGUU---UAAUGCAGACGCAGACUGUGCGUCAUCGU-UUUUCUUG ------------------------(((((((((((.....)))---)..))).((((((.....))))))..)))-)....... ( -15.10, z-score = -1.23, R) >droMoj3.scaffold_6500 15735223 56 - 32352404 ------------------------AAAUGCAGAAAUGAAUAUU---UAAUGCAGACGCAGACGAUGCGUCAUCGU-UUUUCUUG ------------------------...((((..((.......)---)..))))((((((.....)))))).....-........ ( -12.70, z-score = -1.09, R) >droGri2.scaffold_15126 987435 57 + 8399593 ------------------------AAUGGCAAAAAUGCAUAUU---UAAUGCAGACGCAGACGAUGCGUCAUCGUGUUUUGCUG ------------------------...(((((((.(((((...---..)))))((((((.....))))))......))))))). ( -20.70, z-score = -3.17, R) >consensus ______________A_AG__AA_UAGCAGAAAAAGUGCAUU_____CAAUGCAGACGCAGACGAUGCGUCAUCGGUUUUUAUA_ ...................................((((..........))))((((((.....)))))).............. ( -9.16 = -9.62 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:13 2011